if (!require("seriation")) install.packages("seriation")Loading required package: seriationlibrary("seriation")
data("Wood")
Wood <- Wood[sample(nrow(Wood)), sample(ncol(Wood))]
dim(Wood)[1] 136 6DT::datatable(round(Wood, 2))A Heatmap uses colored tiles to represent the values in a data matrix. Patterns can be easily seen if the rows and columns are appropriately reordered. There are many ways to reorder a matrix and the order has a huge impact on how useful the visualization is. The package seriation implements a large number of reordering methods (see: the list with all implemented seriation methods). seriation also provides a set of functions to display reordered heatmaps:
pimage()hmap()gghmap()How to cite the seriation package:
Hahsler M, Hornik K, Buchta C (2008). “Getting things in order: An introduction to the R package seriation.” Journal of Statistical Software, 25(3), 1-34. ISSN 1548-7660, doi:10.18637/jss.v025.i03 https://doi.org/10.18637/jss.v025.i03.
As an example, we use the Wood dataset with the normalized gene expression data (a sample of 136 genes) for wood formation in poplar trees in 6 locations. In case the data has alread some order, we randomly reorder rows and columns for this example.
if (!require("seriation")) install.packages("seriation")Loading required package: seriationlibrary("seriation")
data("Wood")
Wood <- Wood[sample(nrow(Wood)), sample(ncol(Wood))]
dim(Wood)[1] 136 6DT::datatable(round(Wood, 2))Here is a simple heatmap without reordering. No structure is visible.
pimage(Wood)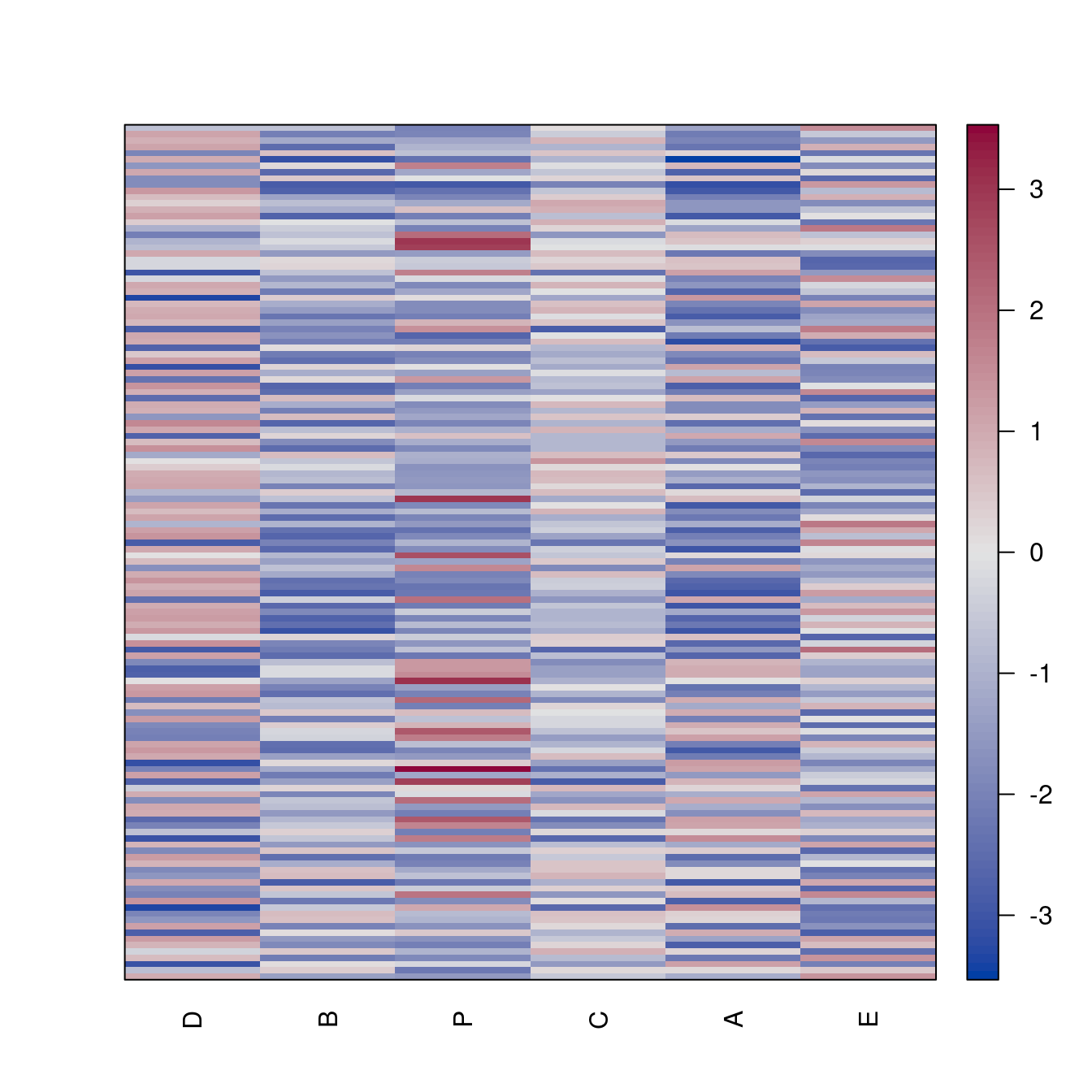
Many seriation methods are available. The manual page for seriate() describes the methods available in package seriation.
Methods of interest for heatmaps are dendrogram leaf order-based methods applied to rows and columns. This is done using method = "heatmap". The actual seriation method can be passed on as parameter seriaton_method, but it has a suitable default if it is omitted. Here is an example:
o <- seriate(Wood, method = "Heatmap", seriation_method = "HC_Mean")
oobject of class 'ser_permutation', 'list'
contains permutation vectors for 2-mode data
vector length seriation method
1 136 Heatmap
2 6 HeatmapThis is the order for rows and columns. The method heatmap automatically performs hierarchical clustering and then applies the seriation method for reordering of dendrogram leafs. Here we use the row/column mean to reorder the dendrogram. The resulting order (2 means second dimension, i.e., columns) can be shown, and the reordered dendrogram and a reorderd image can be plotted.
get_order(o, 2)B A P E C D
2 5 1 3 6 4 plot(o[[2]])
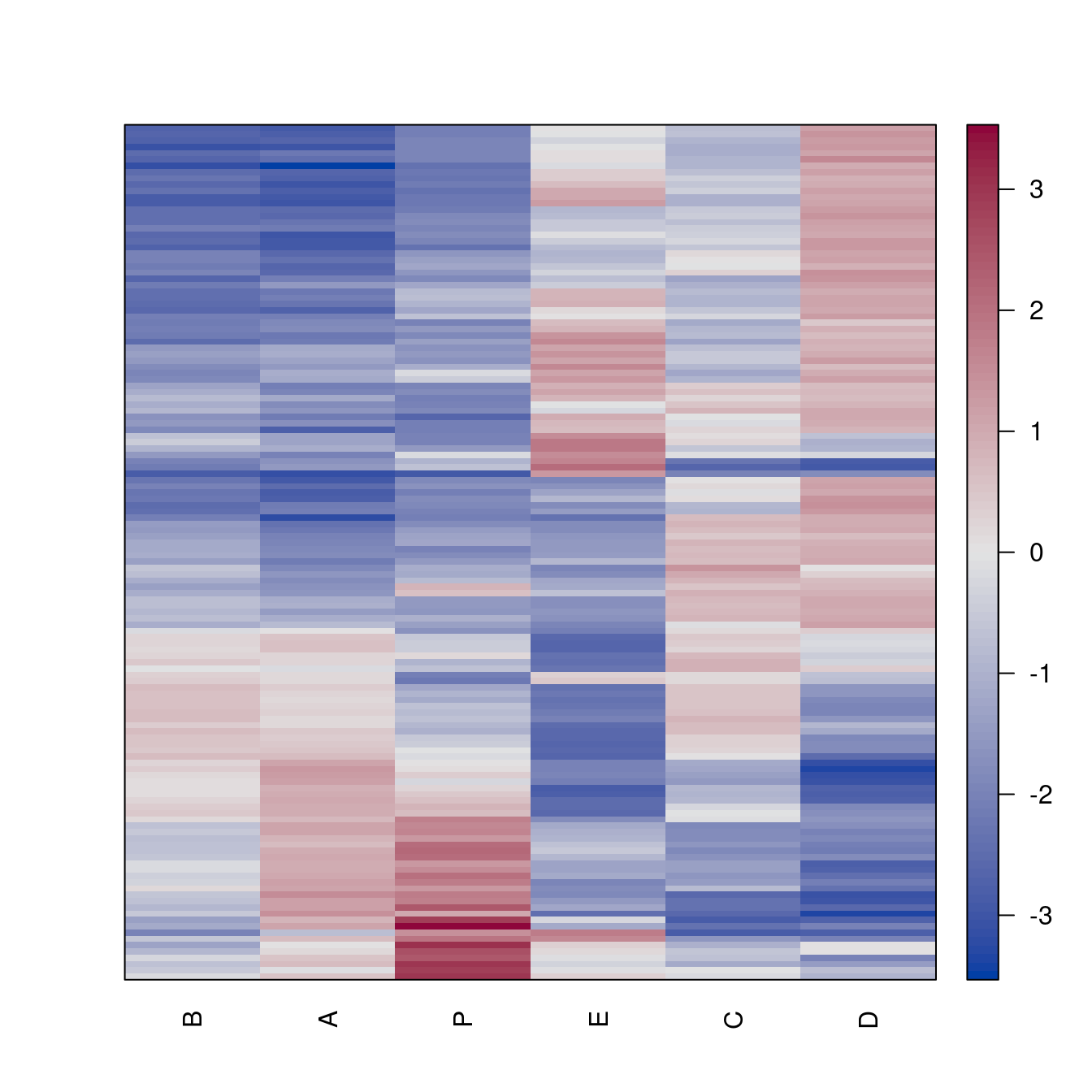
pimage(Wood, order = o)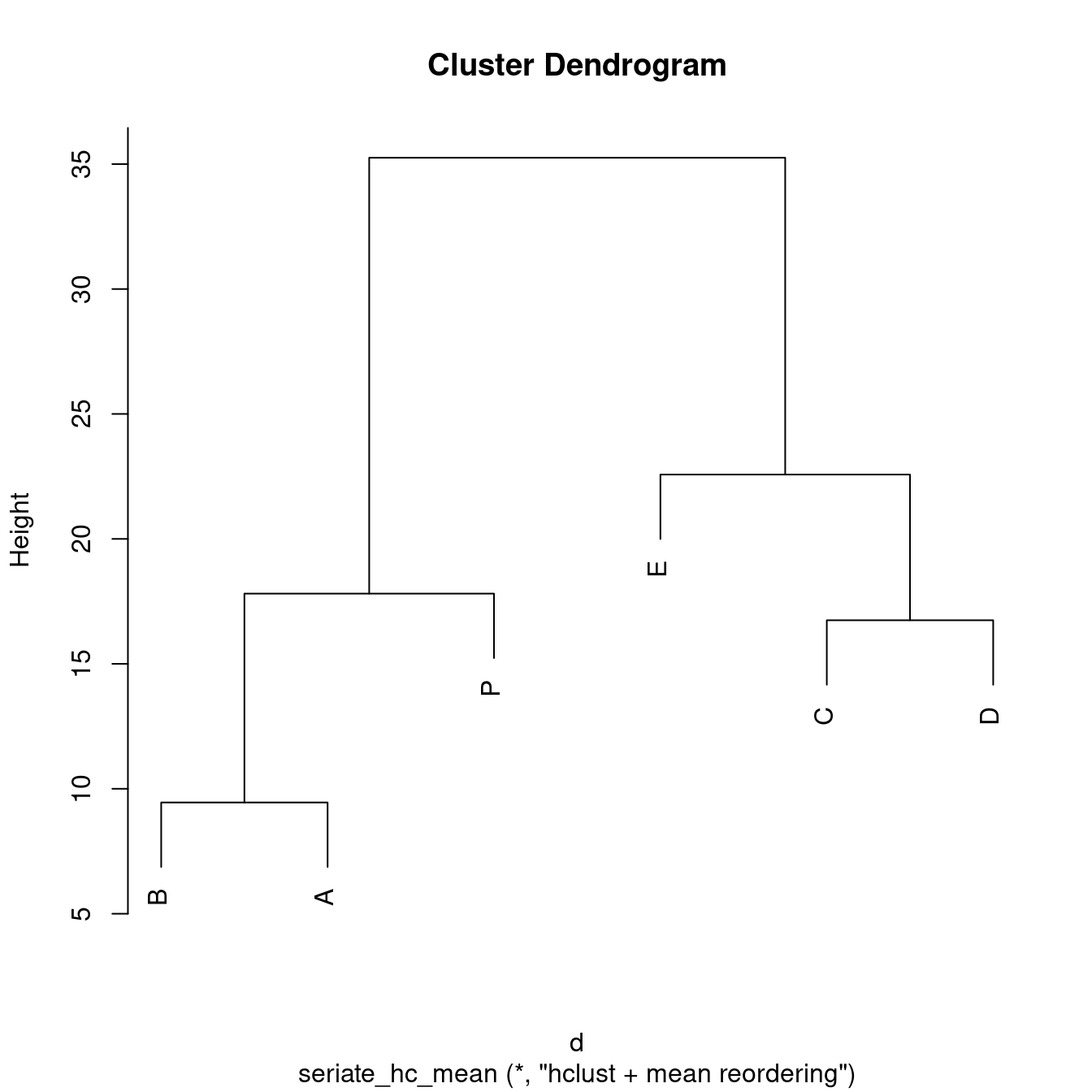
Package seriation has several functions to display heatmaps.
The permutation image plot in seriation provides a simple heatmap. The order argument not only accepts a seriation order, but also a seriation method.
pimage(Wood, order = "Heatmap", seriation_method = "HC_complete",
main = "Wood (hierarichal clustering)")
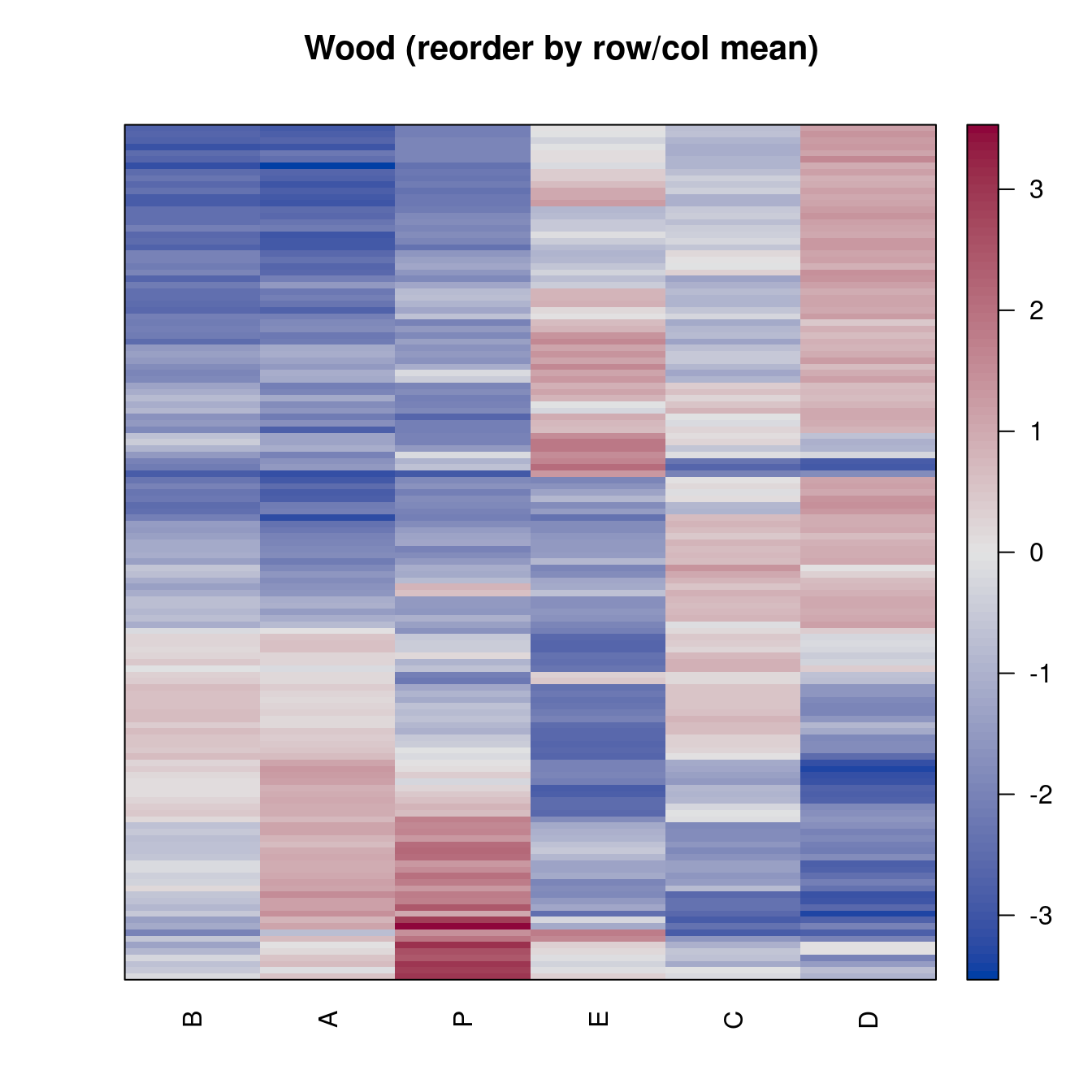
pimage(Wood, order = "Heatmap", seriation_method = "HC_Mean",
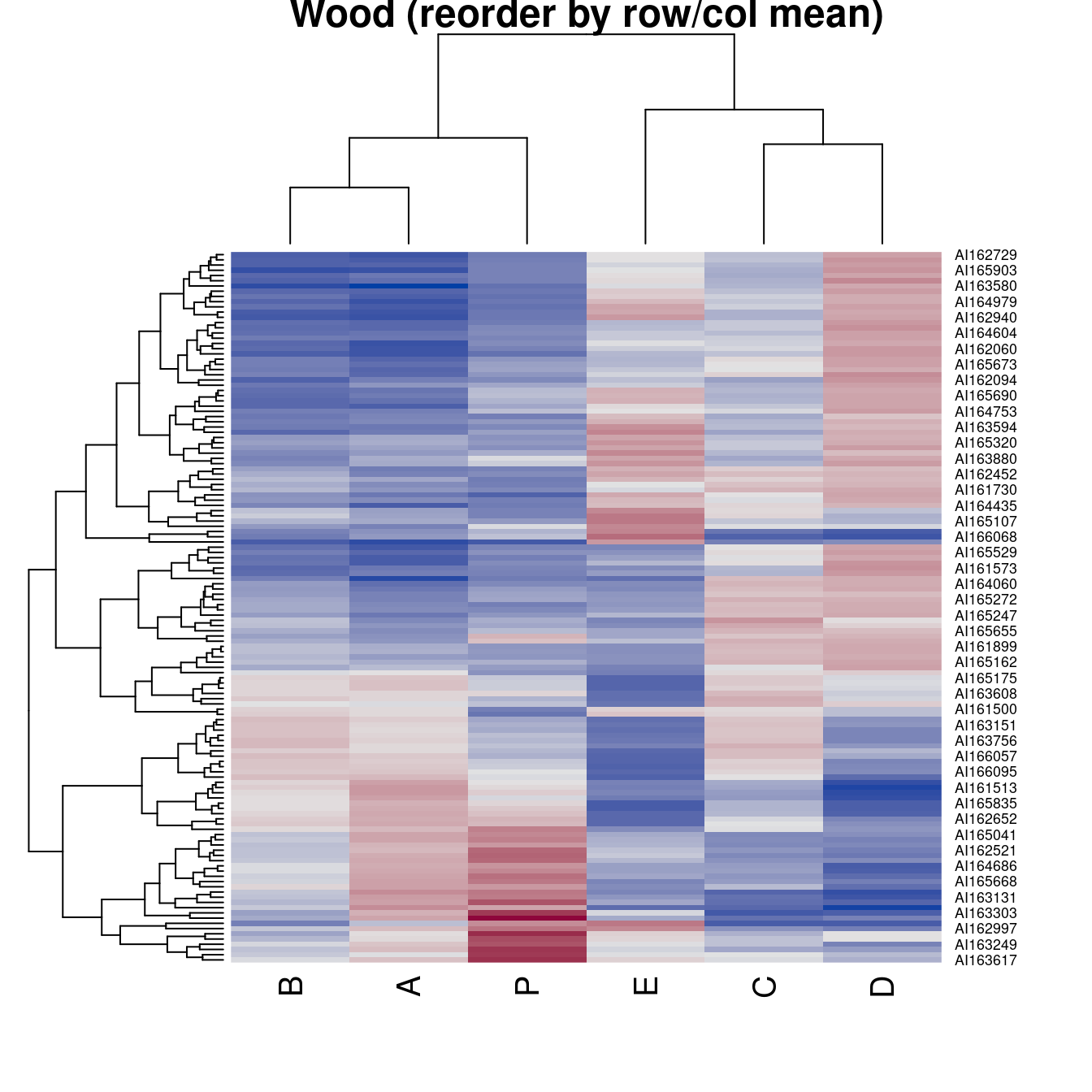
main = "Wood (reorder by row/col mean)")
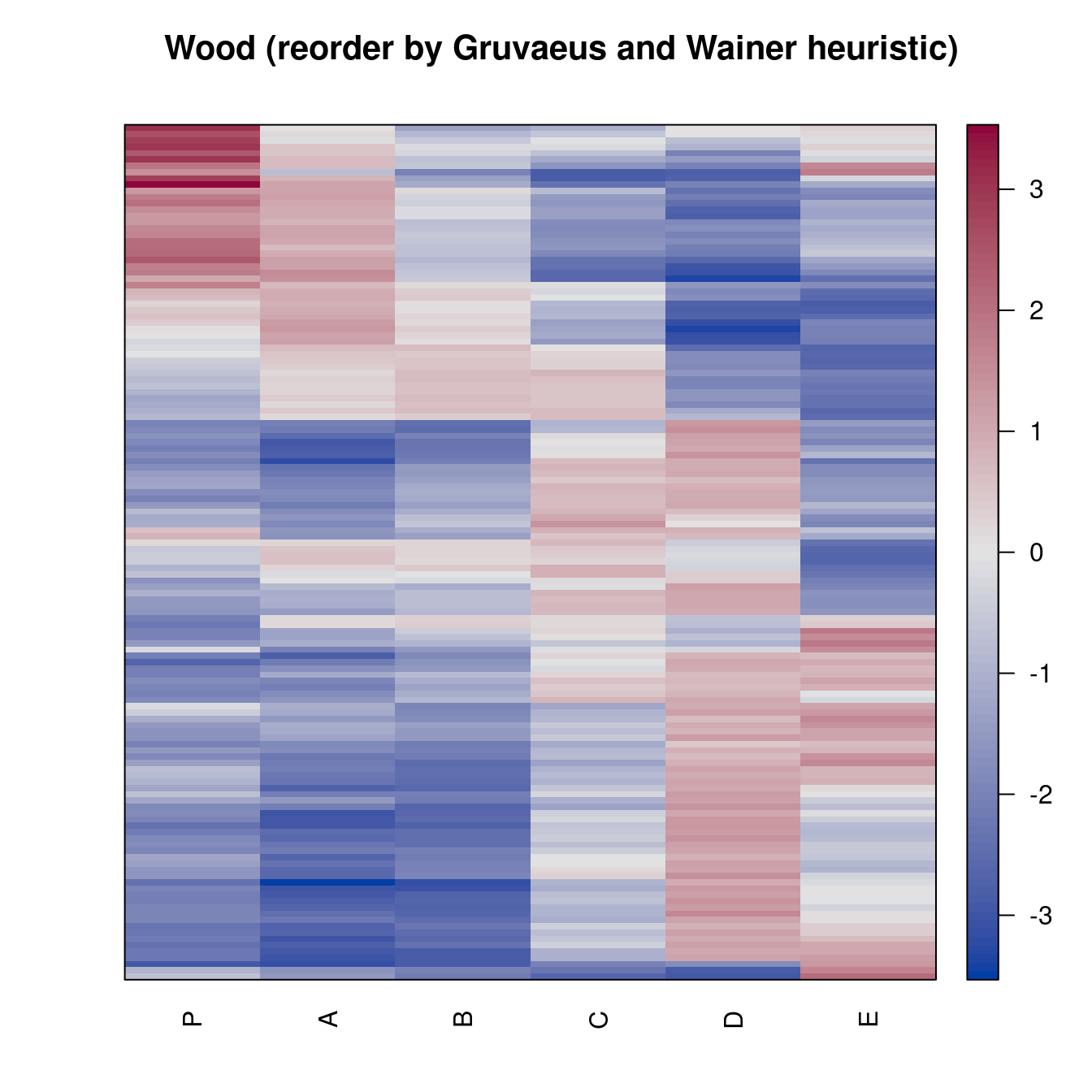
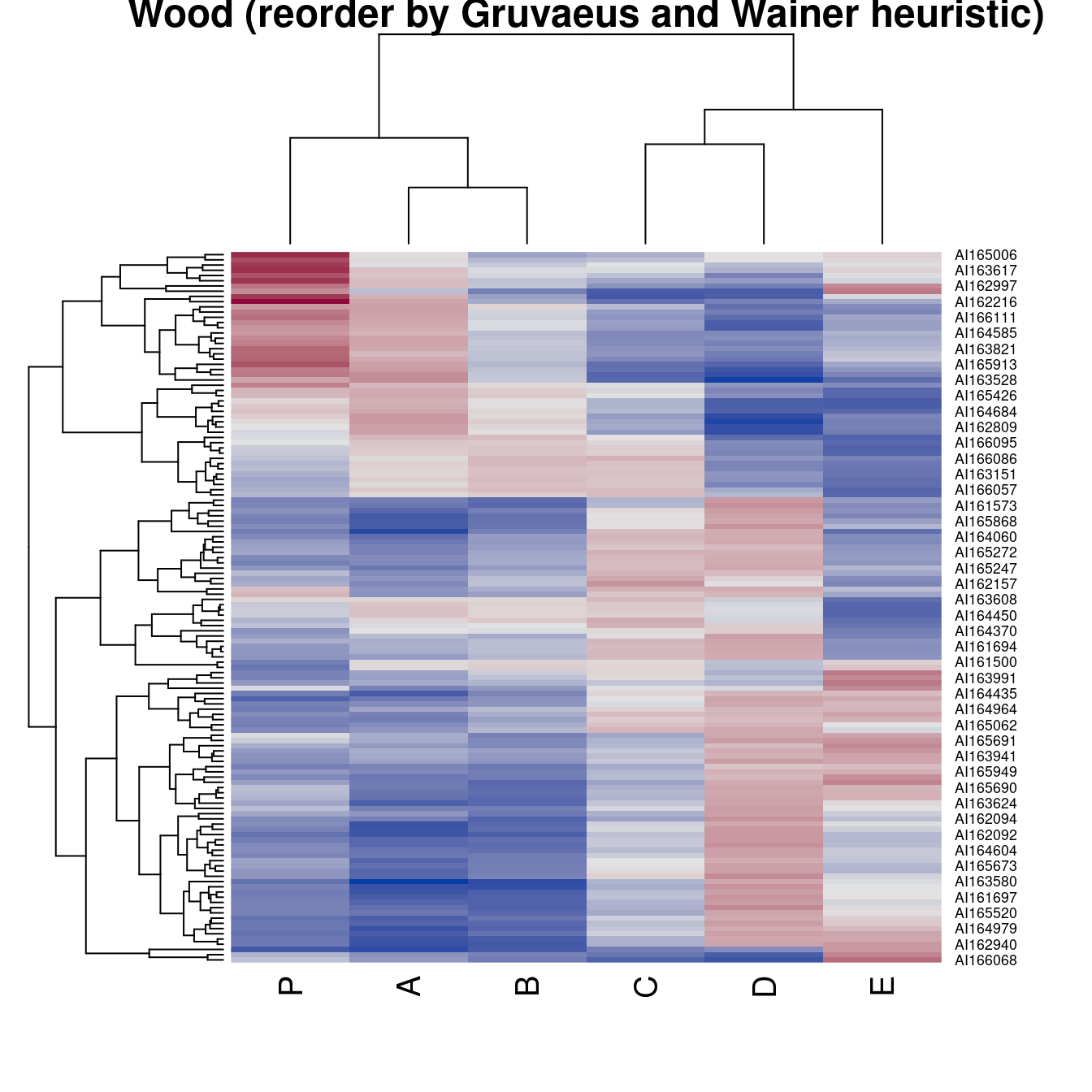
pimage(Wood, order = "Heatmap", seriation_method = "GW_complete",
main = "Wood (reorder by Gruvaeus and Wainer heuristic)")Registered S3 method overwritten by 'gclus':
method from
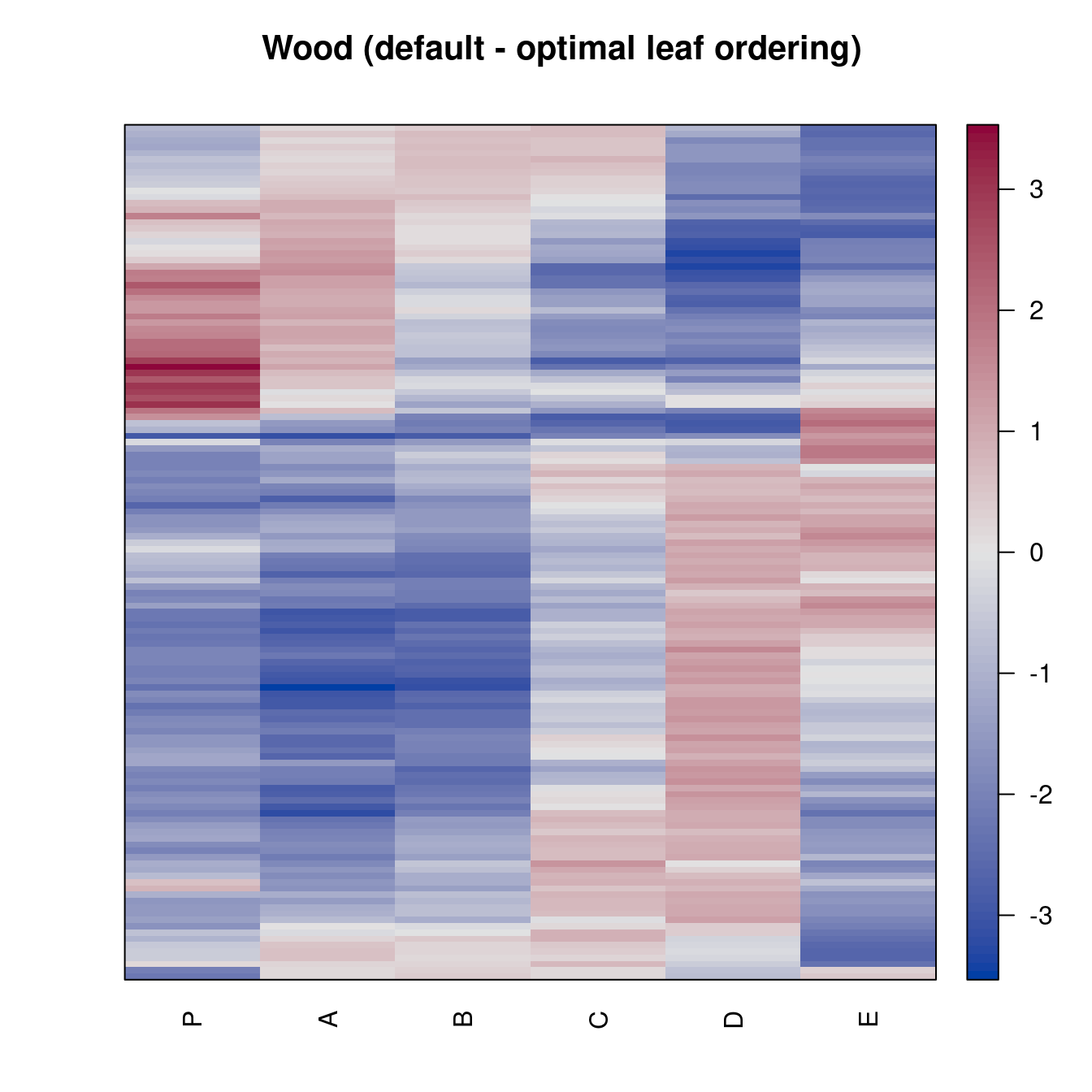
reorder.hclust seriationpimage(Wood, order = "Heatmap",
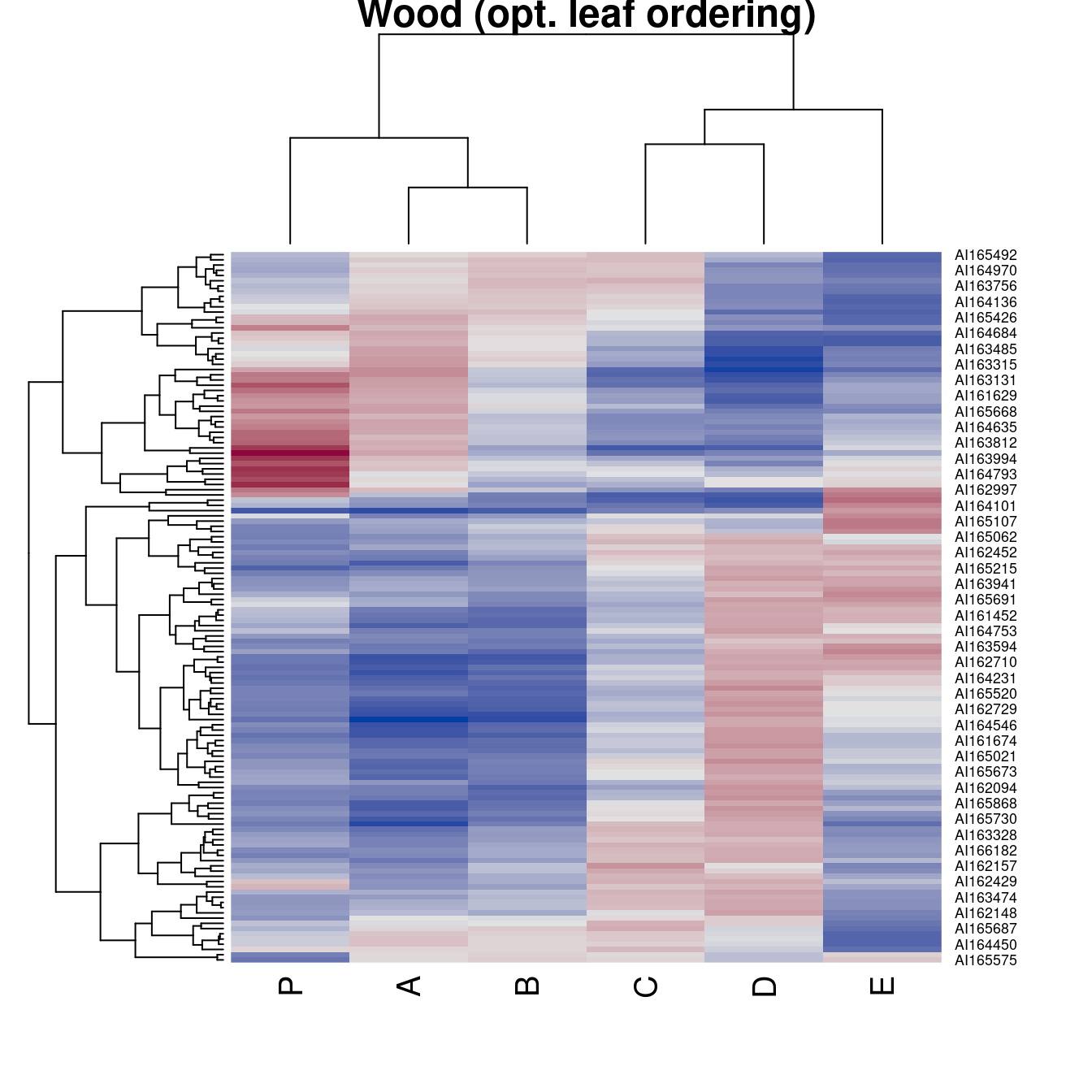
main = "Wood (default - optimal leaf ordering)")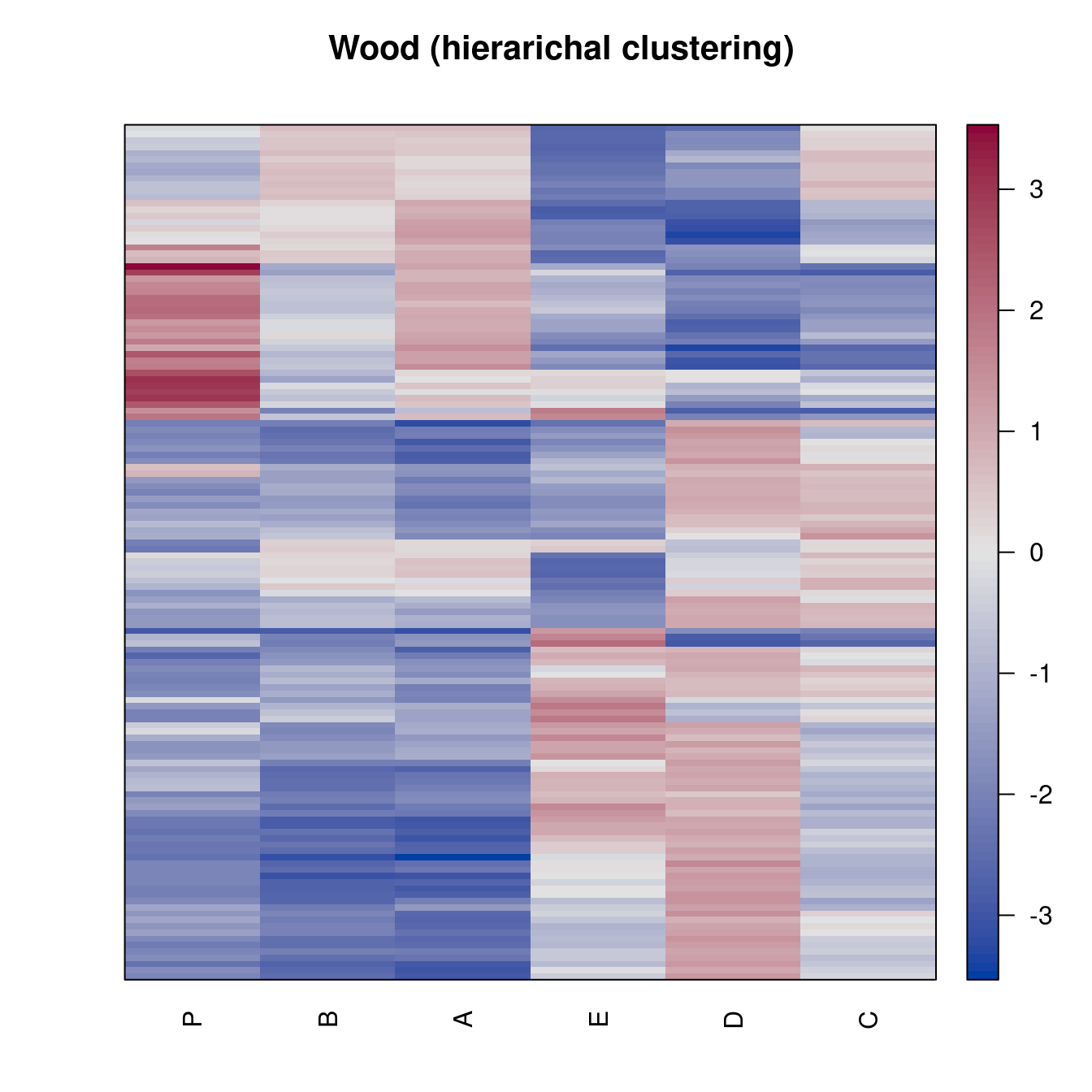
Here are some typical reordering schemes.
hmap(Wood, method = "HC_complete", main = "Wood (hierarichal clustering)")
hmap(Wood, method = "HC_Mean", main = "Wood (reorder by row/col mean)")
hmap(Wood, method = "GW_complete", main = "Wood (reorder by Gruvaeus and Wainer heuristic)")
hmap(Wood, method = "OLO_complete", main = "Wood (opt. leaf ordering)")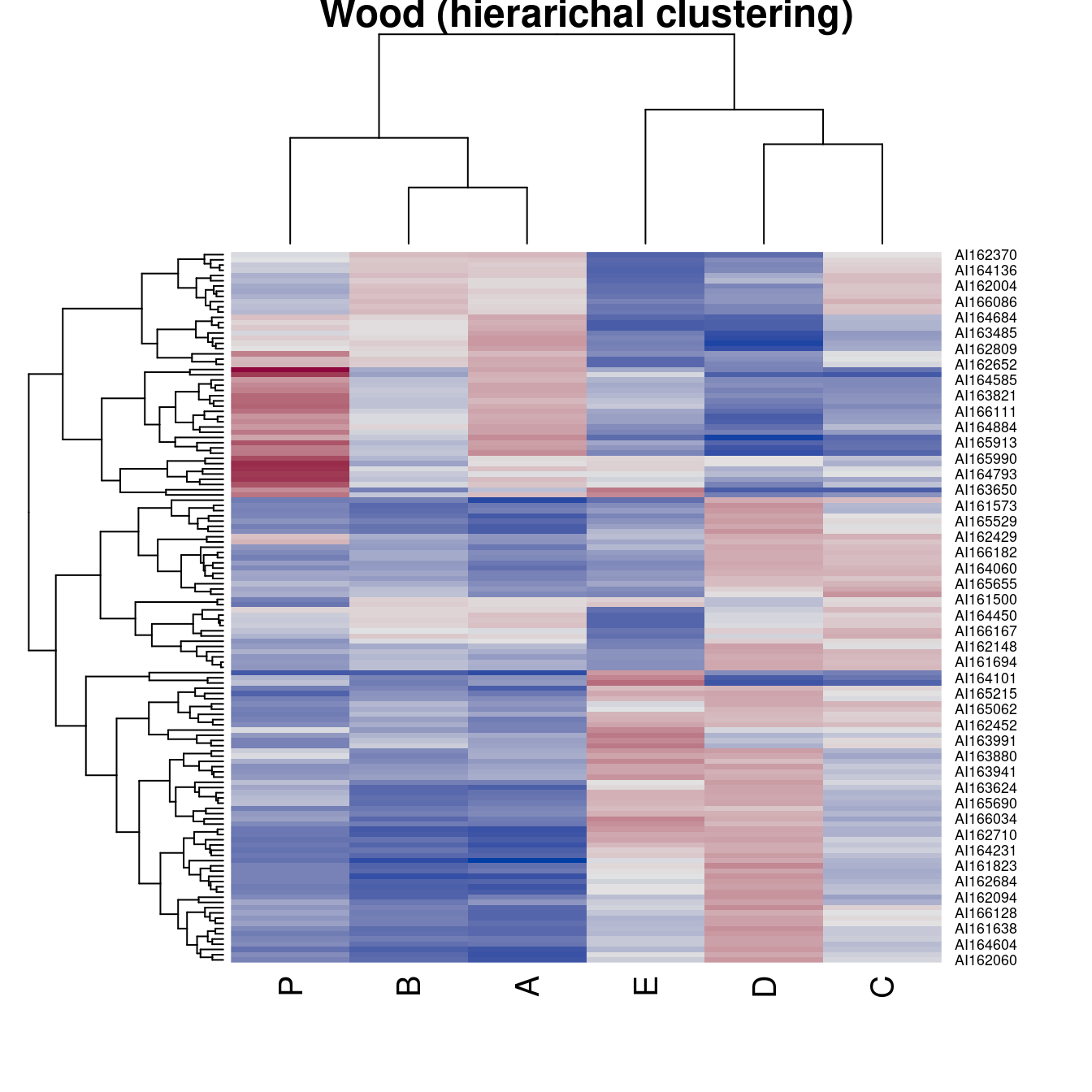
Different linkage types can be added in the method name.
Package DendSer offers more dendrogram seriation methods. These methods can be registered using `register_DendSer()``
register_DendSer()Registering new seriation method 'DendSer' for 'dist'register_DendSer()Registering new seriation method 'DendSer_BAR' for 'dist'register_DendSer()Registering new seriation method 'DendSer_PL' for 'dist'register_DendSer()Registering new seriation method 'DendSer_LPL' for 'dist'register_DendSer()Registering new seriation method 'DendSer_ARc' for 'dist'register_DendSer()Registering new seriation criteron 'ARc' for 'dist' using register_DendSer()hmap(Wood, method = "DendSer_BAR", main = "Wood (banded anti-Robinson)")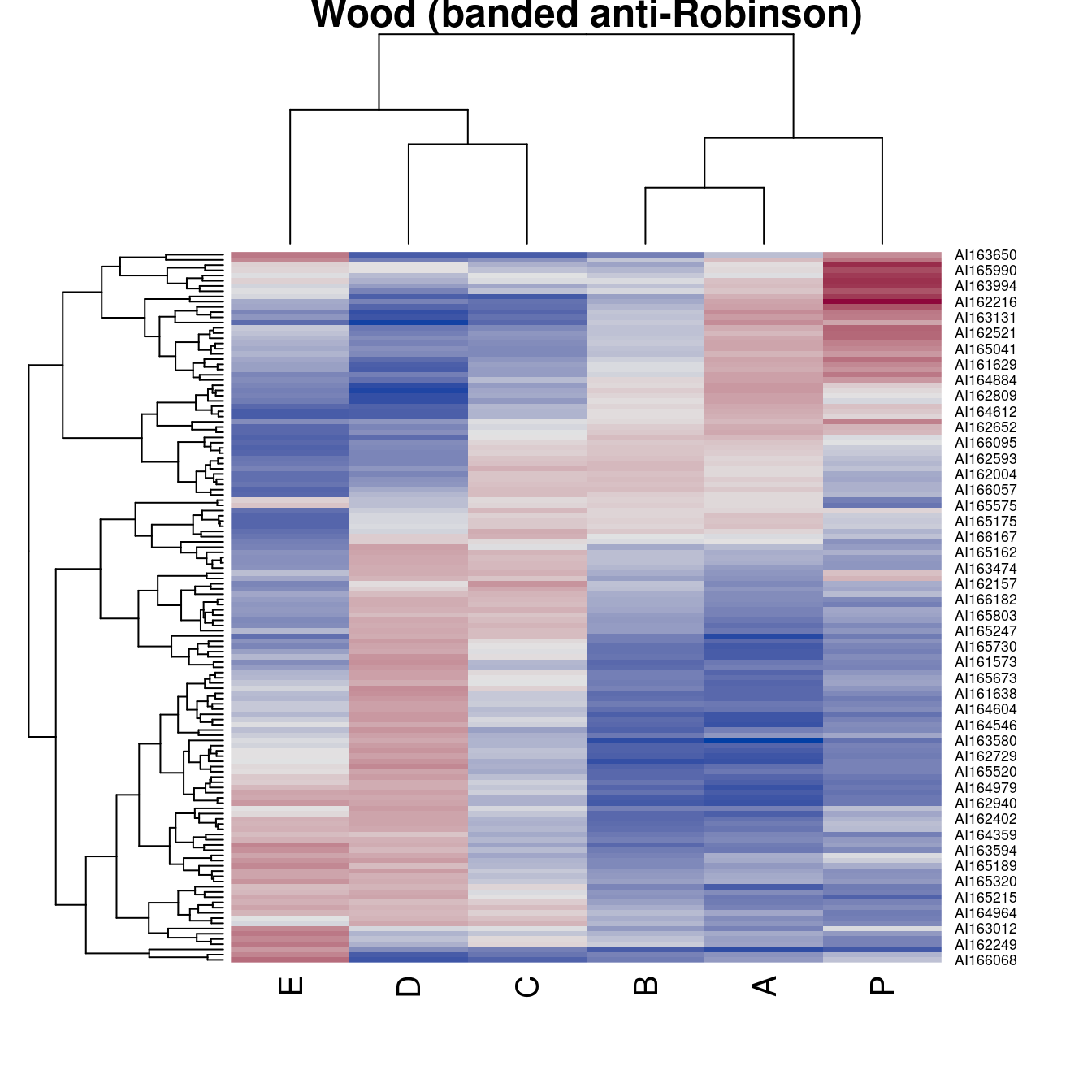
Instead of dendrograms, also reordered distance matrices can be displayed. Dark block around the diagonal indicate the cluster structure.
hmap(Wood, method = "HC_complete",
plot_margins = "distances",
main = "Wood (hierarichal clustering)")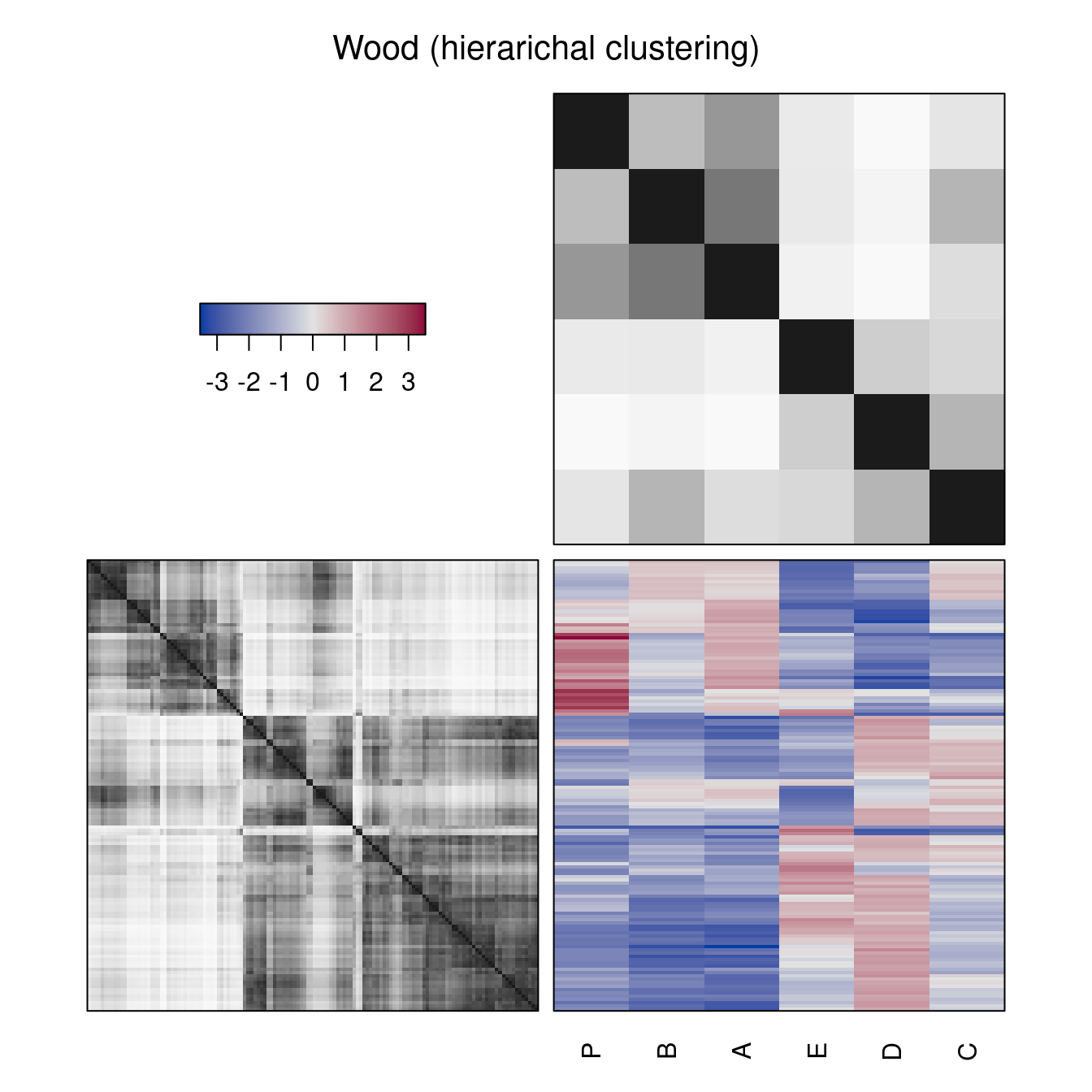
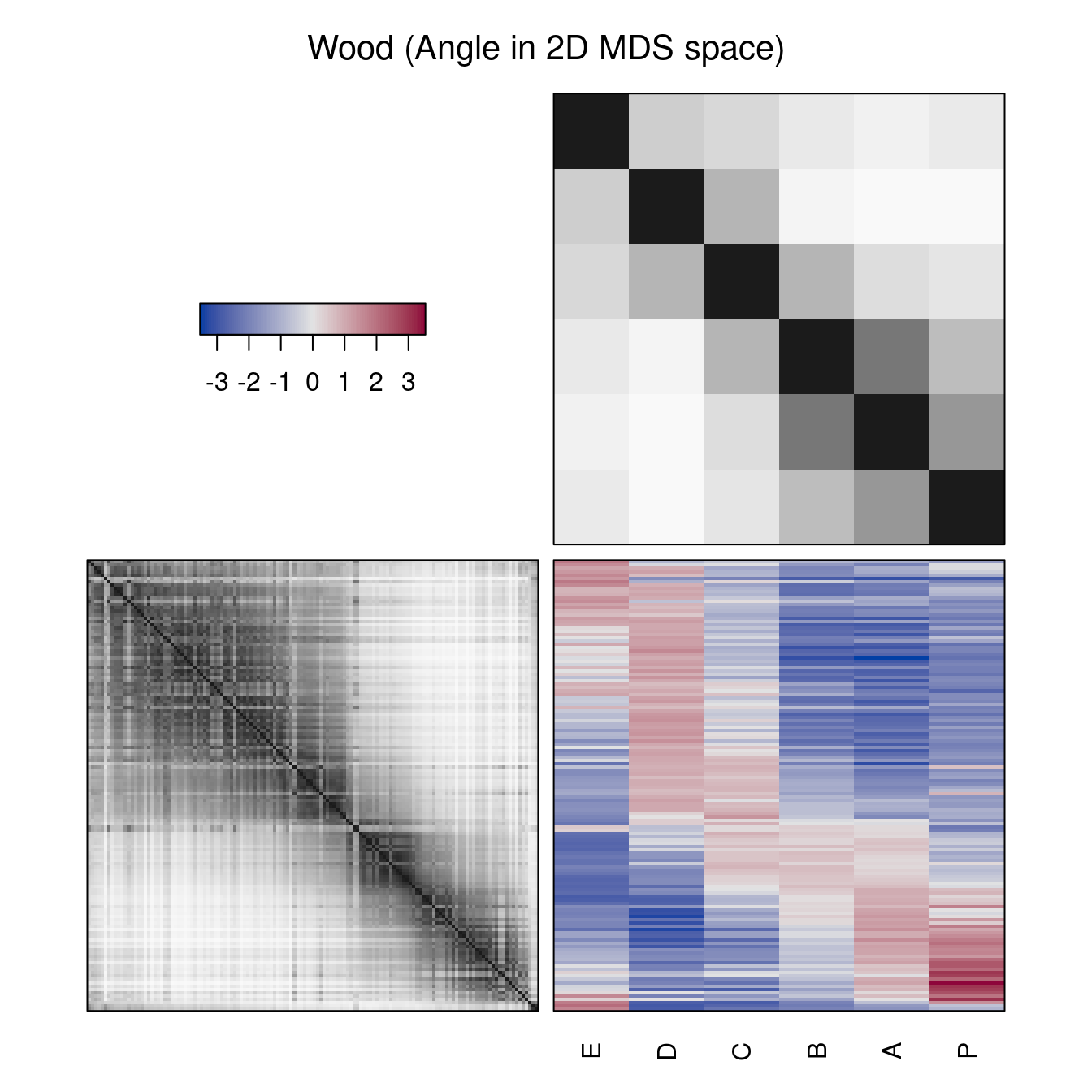
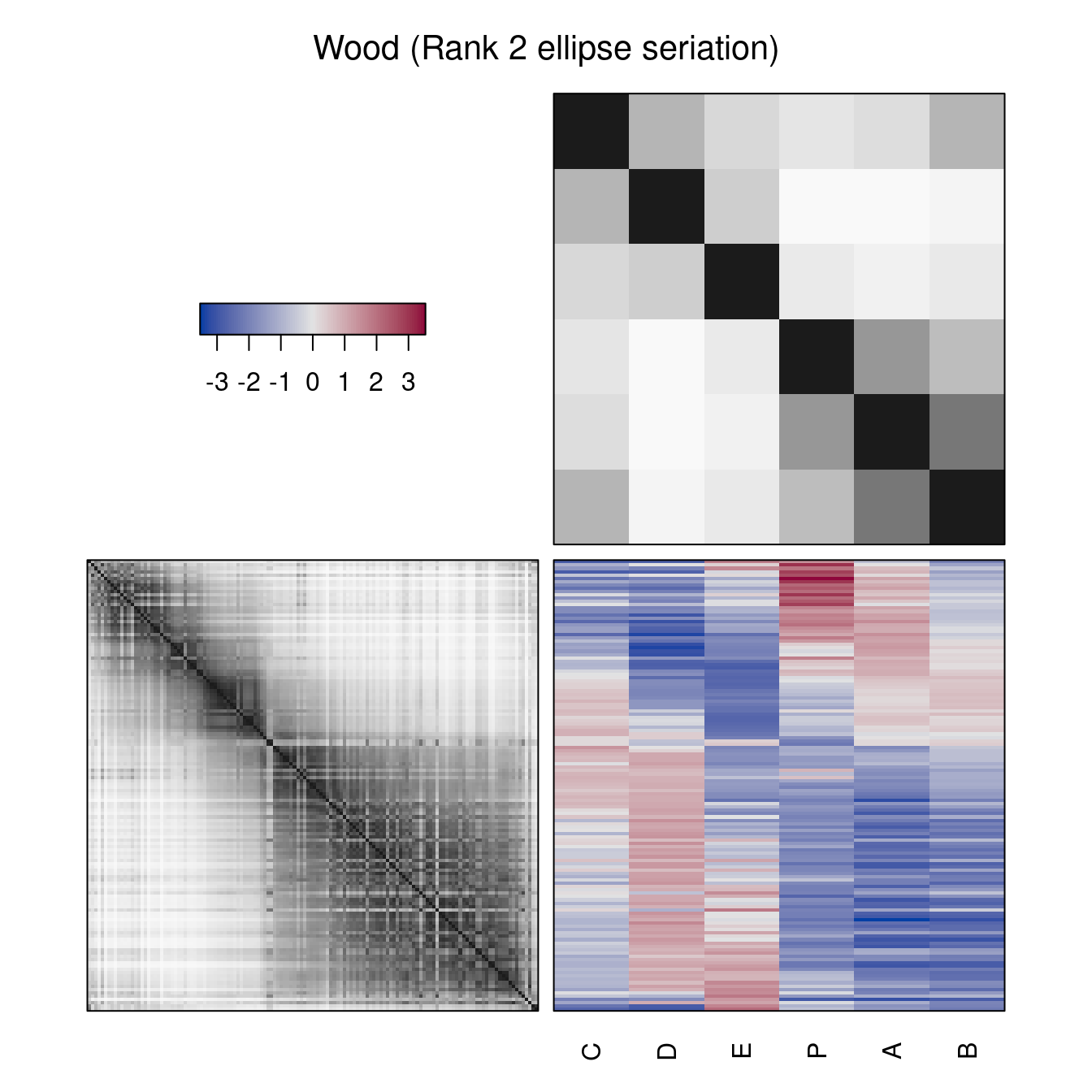
Also non-dendrogram-based reordering methods can be used. These methods reorder rows and columns. Instead of the dendrograms, reordered distance matrices are shown.
hmap(Wood, method = "MDS", main = "Wood (MDS)")
hmap(Wood, method = "MDS_angle", main = "Wood (Angle in 2D MDS space)")
hmap(Wood, method = "R2E", main = "Wood (Rank 2 ellipse seriation)")
hmap(Wood, method = "TSP", main = "Wood (Traveling salesperson)")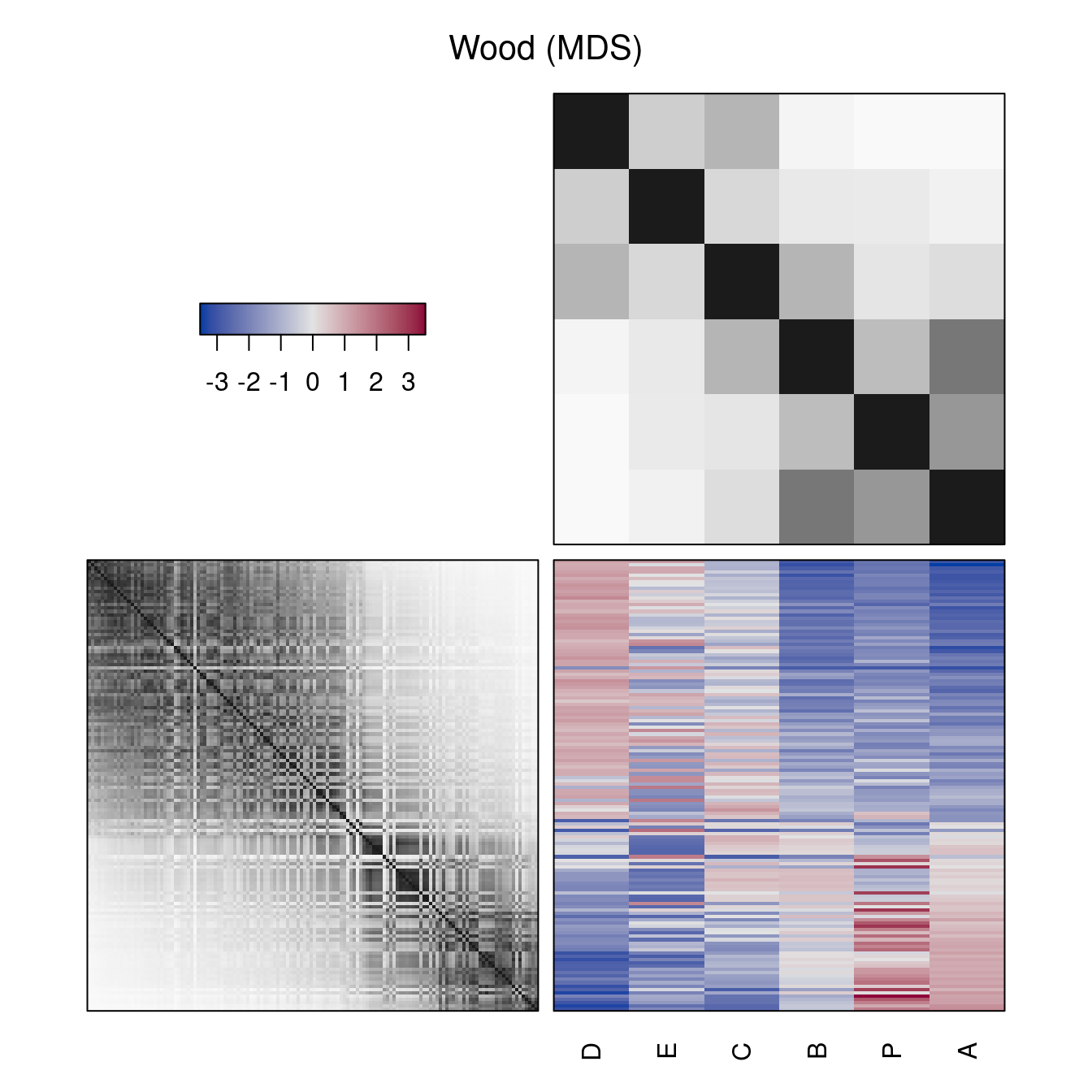
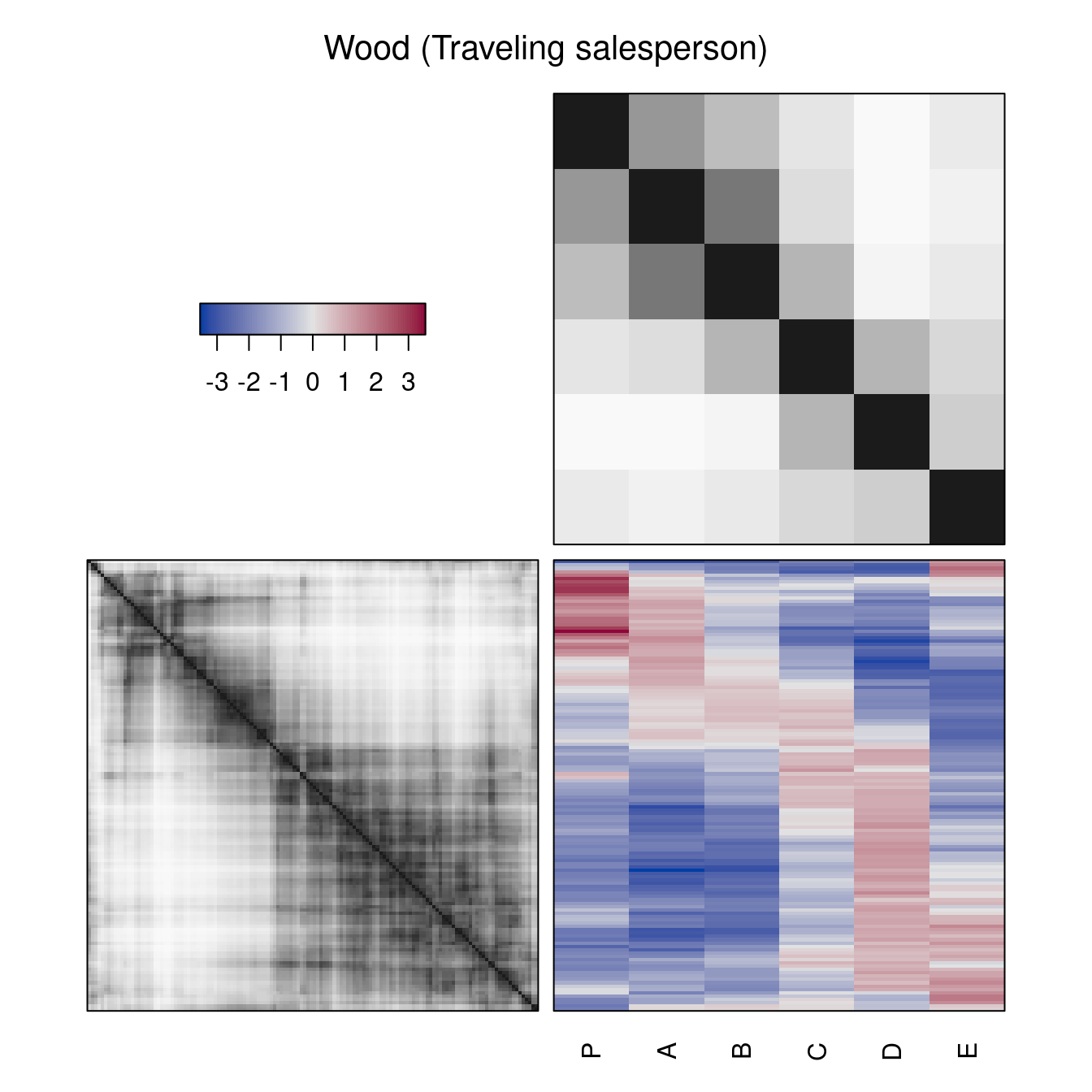
hmap(Wood, col = grays())
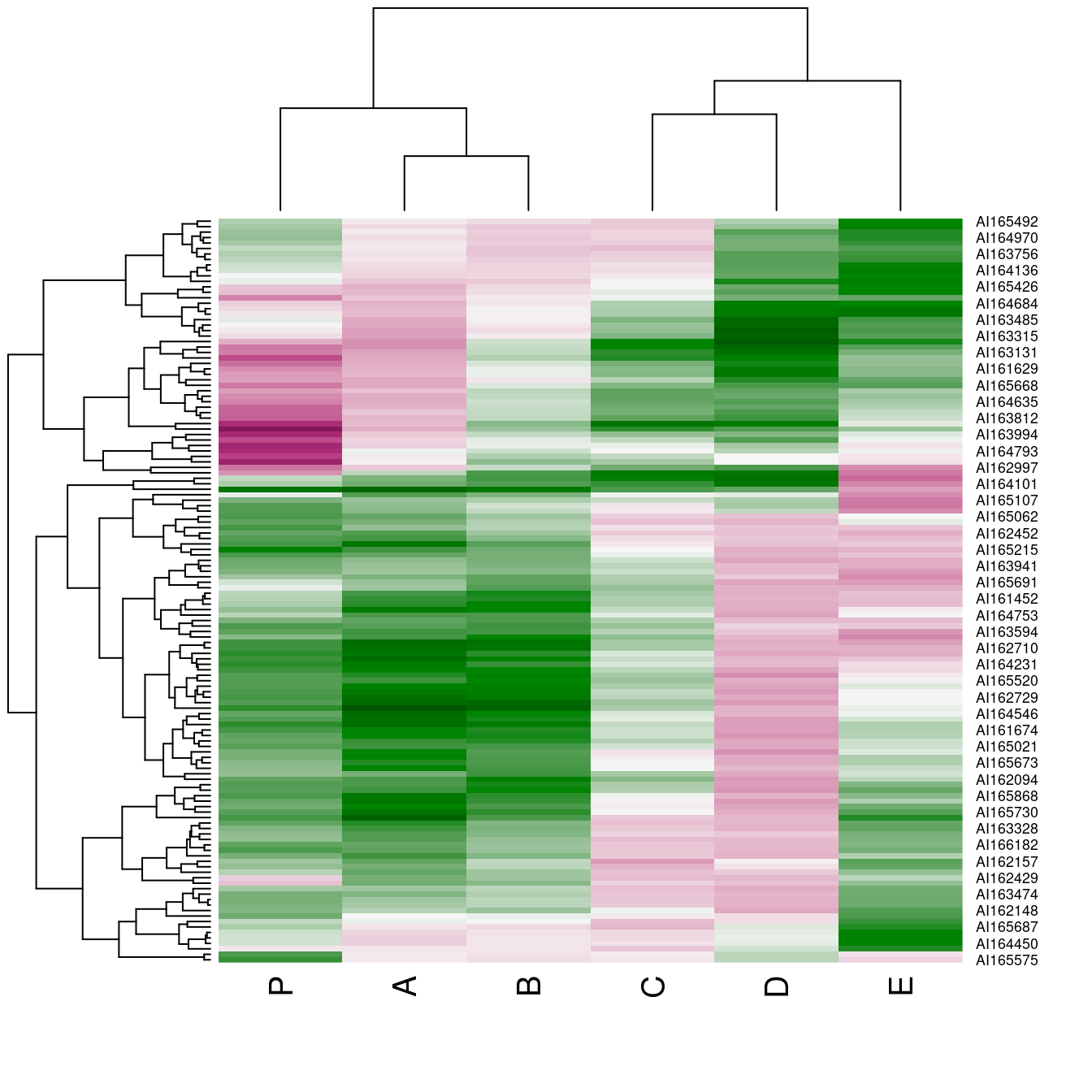
hmap(Wood, col = greenred())
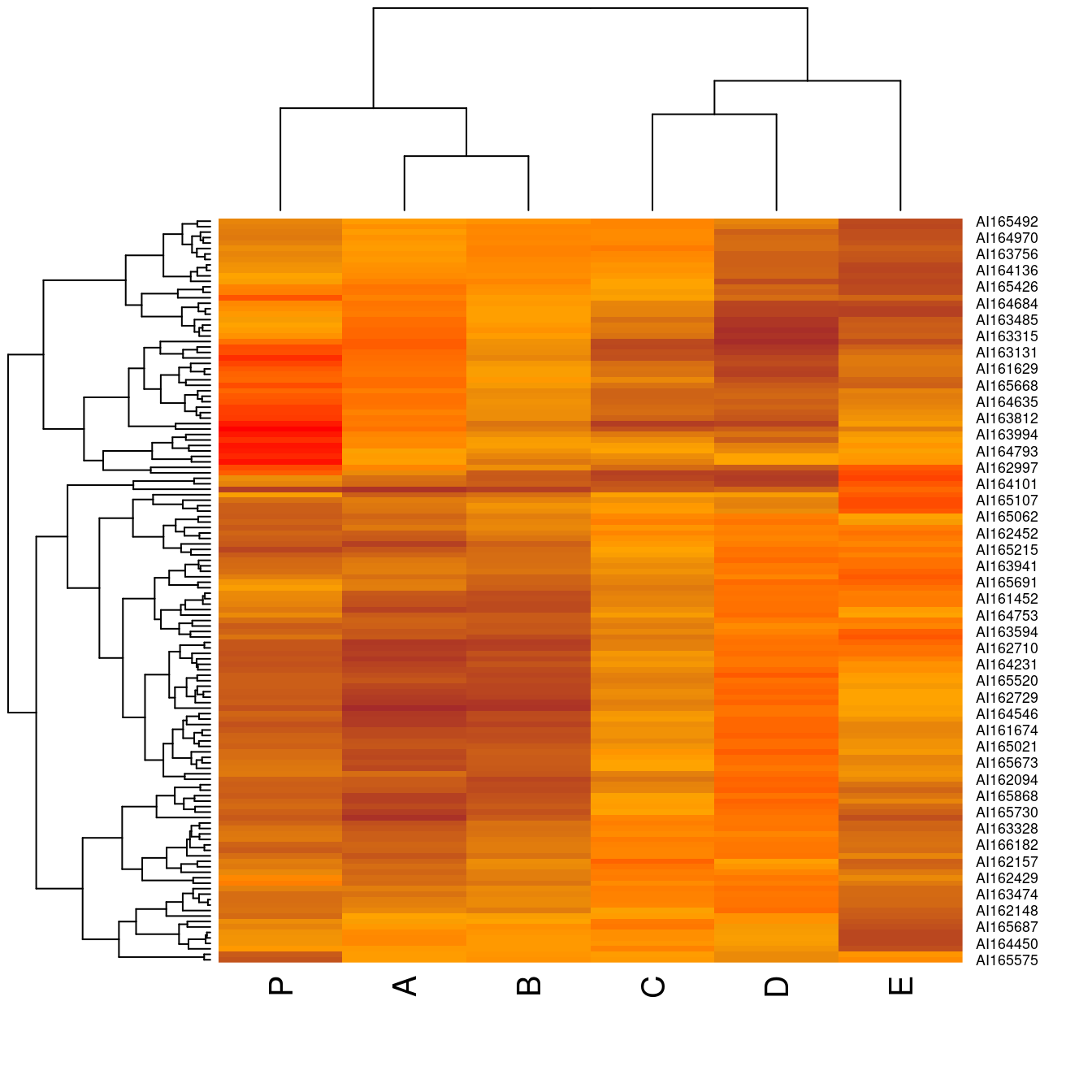
hmap(Wood, col = colorRampPalette(c("brown", "orange", "red"))( 100 ) )
if (!require("viridis")) install.packages("viridis")Loading required package: viridisLoading required package: viridisLitehmap(Wood, col = viridis::viridis(100))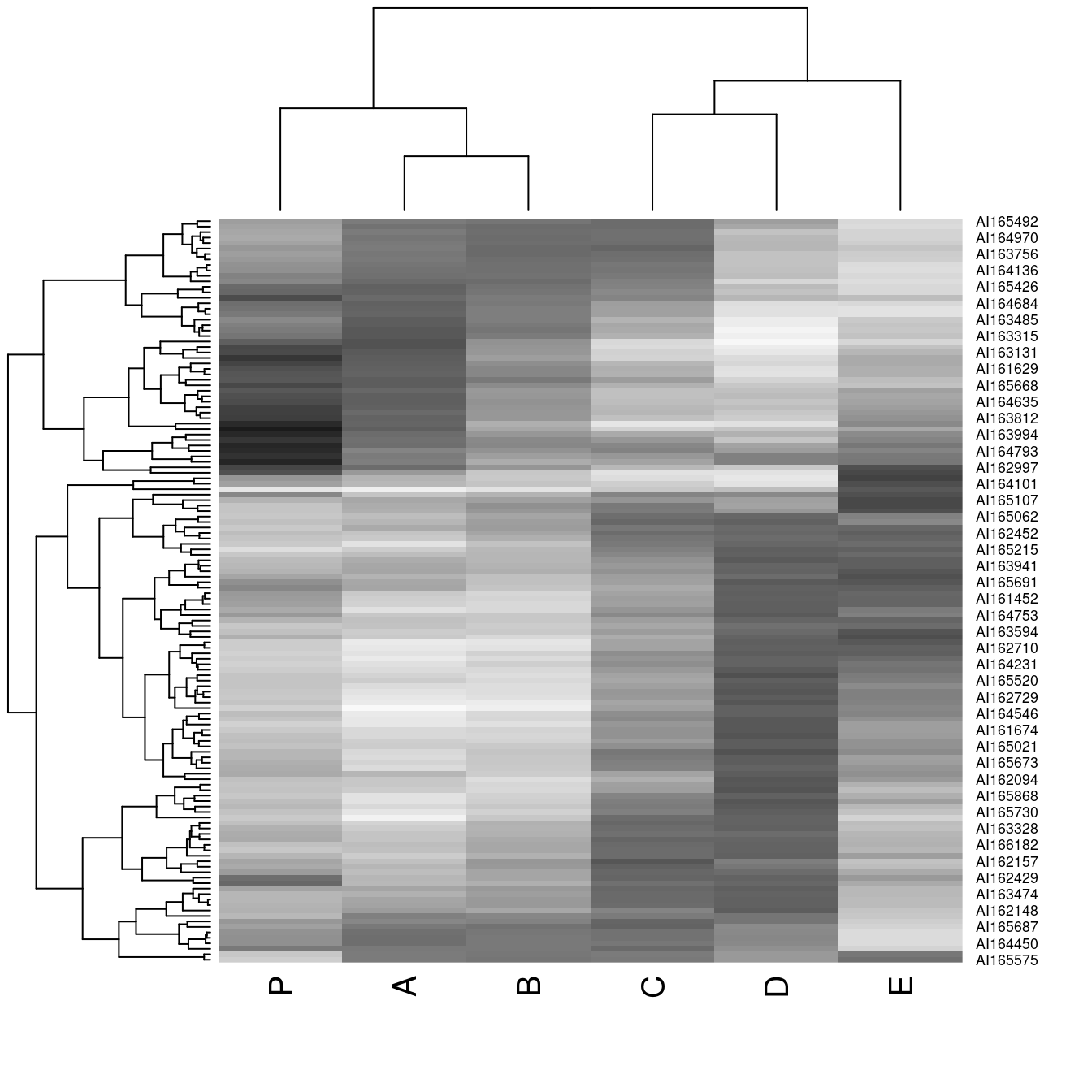
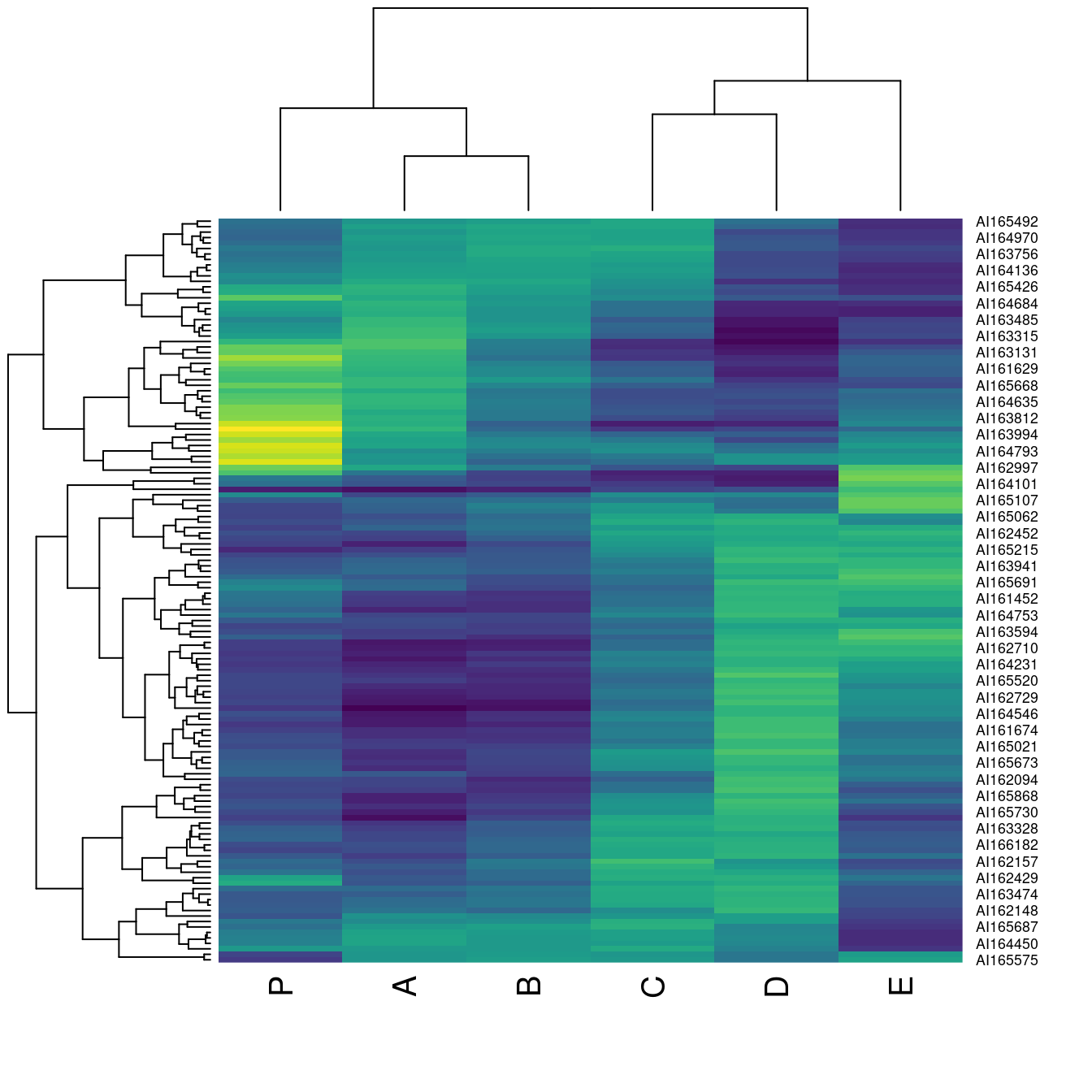
There are many other packages to create color palettes in R like RColorBrewer or colorspaces.
All options are also available for ggplot2 using gghmap(). Currently there is no support to display dendrograms.
if (!require("ggplot2")) install.packages("ggplot2")Loading required package: ggplot2library(ggplot2)
gghmap(Wood, method = "OLO")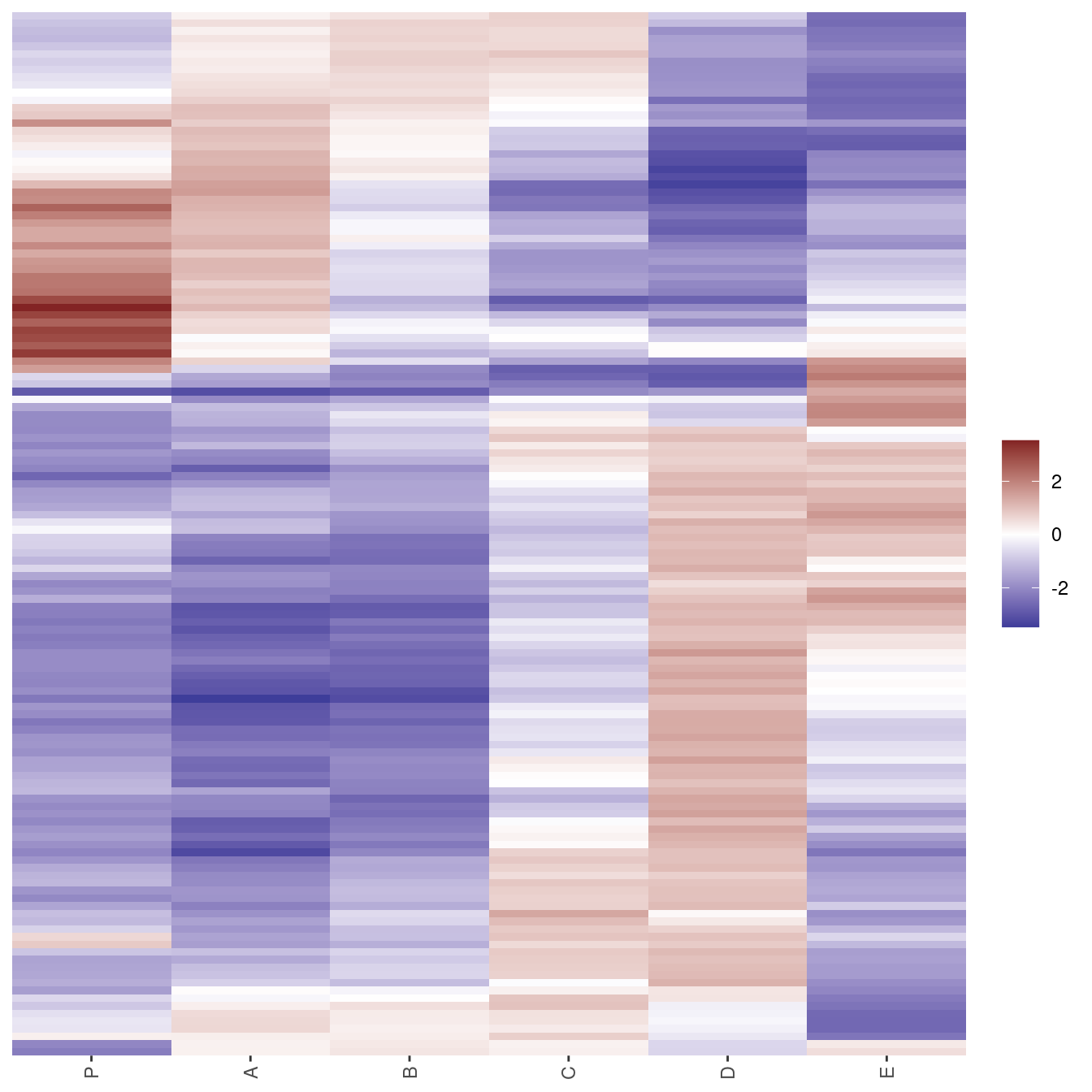
The package seriation can be used to compute reordering for other heatmap packages.
This is R’s standard heatmap function. seriate() can be used to supply the reordered dendrogram.
o <- seriate(Wood, method = "Heatmap", seriation_method = "OLO")
heatmap(Wood, Rowv = as.dendrogram(o[[1]]), Colv = as.dendrogram(o[[2]]))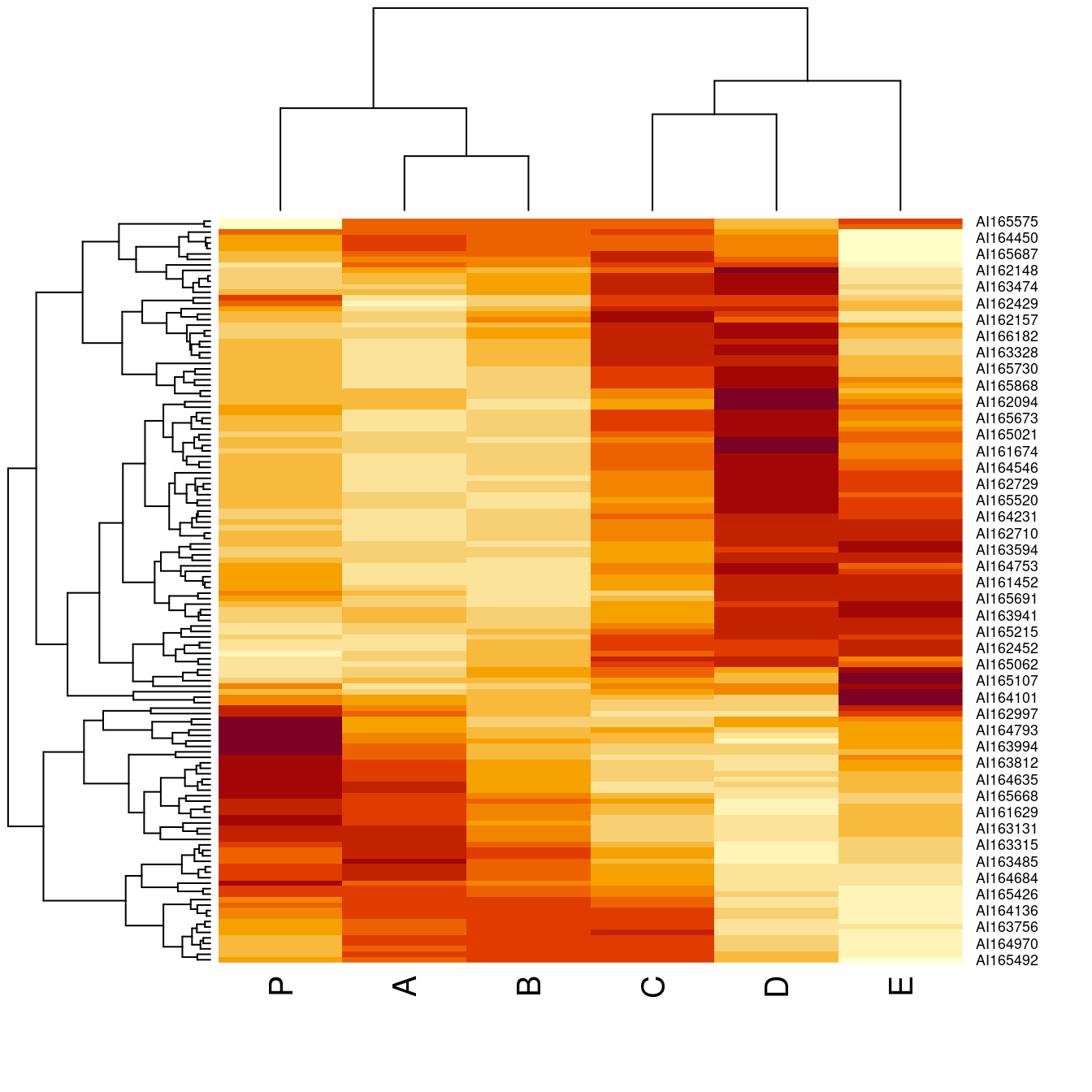
We can also supply the rank order for any seriation method as weights and the dendrogram will be reordered as close as possible to the seriation order.
o <- seriate(Wood, method = "Heatmap", seriation_method = "Spectral")
heatmap(Wood, Rowv = get_rank(o, 1), Colv = get_rank(o, 2))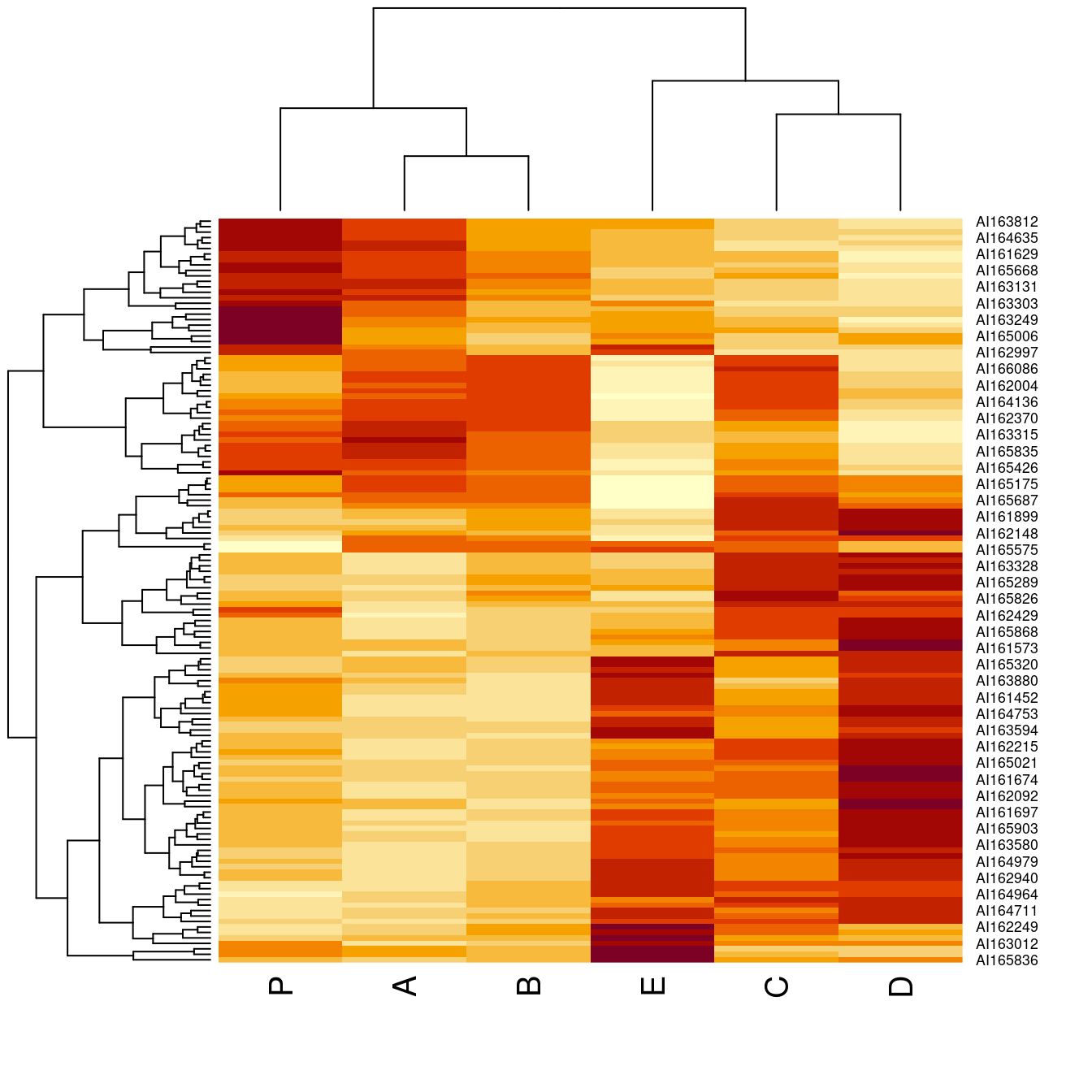
Default ordering using hierarchical clustering.
if (!require("superheat")) install.packages("superheat")Loading required package: superheatlibrary("superheat")
superheat(Wood,
row.dendrogram = TRUE, col.dendrogram = TRUE)Warning in seq.default(min.col, max.col, length = 100): partial argument match
of 'length' to 'length.out'Warning in seq.default(0, 1, length = length(heat.pal)): partial argument match
of 'length' to 'length.out'Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.
ℹ The deprecated feature was likely used in the superheat package.
Please report the issue to the authors.Warning in rep(c("grey95", "grey80"), length = length(names)): partial argument
match of 'length' to 'length.out'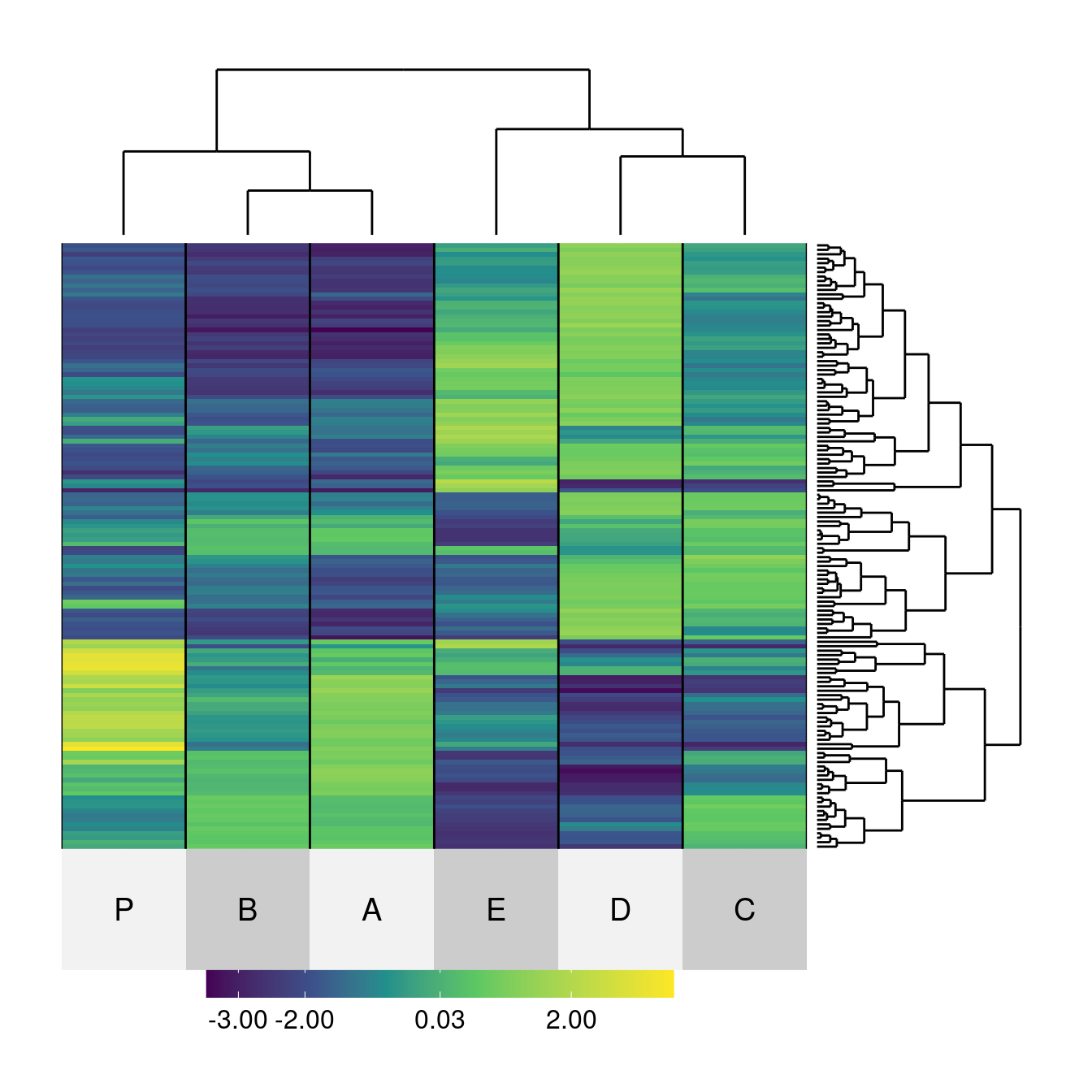
Order with `seriation`. Currently, the dendrograms are not reordered.
o <- seriate(Wood, method = "Heatmap", seriation_method = "OLO_ward")
superheat(Wood,
order.row = as.vector(get_order(o, 1)), order.col = as.vector(get_order(o, 2)),
row.dendrogram = FALSE, col.dendrogram = FALSE)Warning in superheat(Wood, order.row = as.vector(get_order(o, 1)), order.col =
as.vector(get_order(o, : partial argument match of 'order.col' to 'order.cols'Warning in superheat(Wood, order.row = as.vector(get_order(o, 1)), order.col =
as.vector(get_order(o, : partial argument match of 'order.row' to 'order.rows'Warning in seq.default(min.col, max.col, length = 100): partial argument match
of 'length' to 'length.out'Warning in seq.default(0, 1, length = length(heat.pal)): partial argument match
of 'length' to 'length.out'Warning in rep(c("grey95", "grey80"), length = length(names)): partial argument
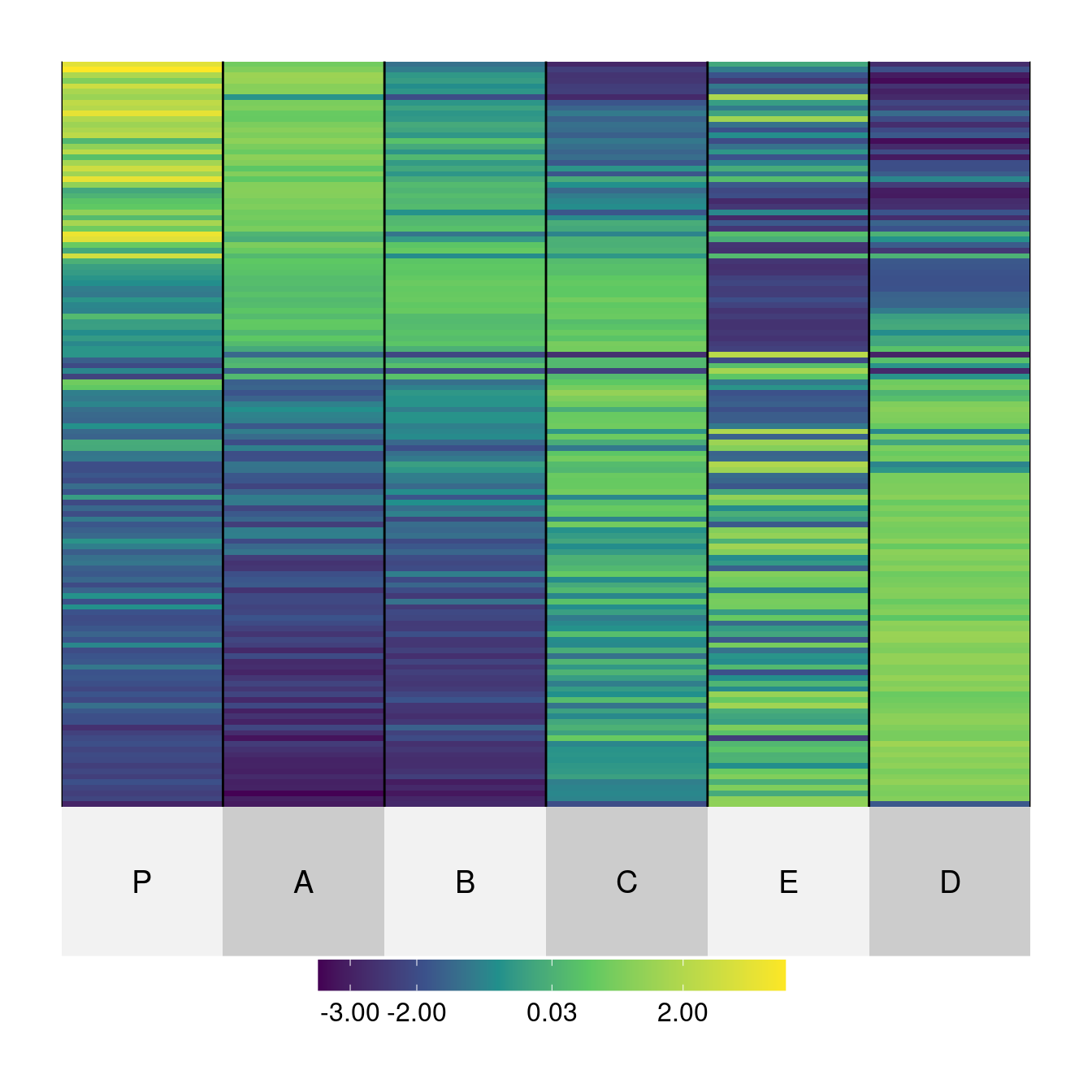
match of 'length' to 'length.out'o <- seriate(Wood, method = "Heatmap", seriation_method = "Spectral")
superheat(Wood,
order.row = as.vector(get_order(o, 1)), order.col = as.vector(get_order(o, 2)),
row.dendrogram = FALSE, col.dendrogram = FALSE)Warning in superheat(Wood, order.row = as.vector(get_order(o, 1)), order.col =
as.vector(get_order(o, : partial argument match of 'order.col' to 'order.cols'Warning in superheat(Wood, order.row = as.vector(get_order(o, 1)), order.col =
as.vector(get_order(o, : partial argument match of 'order.row' to 'order.rows'Warning in seq.default(min.col, max.col, length = 100): partial argument match
of 'length' to 'length.out'Warning in seq.default(0, 1, length = length(heat.pal)): partial argument match
of 'length' to 'length.out'Warning in rep(c("grey95", "grey80"), length = length(names)): partial argument
match of 'length' to 'length.out'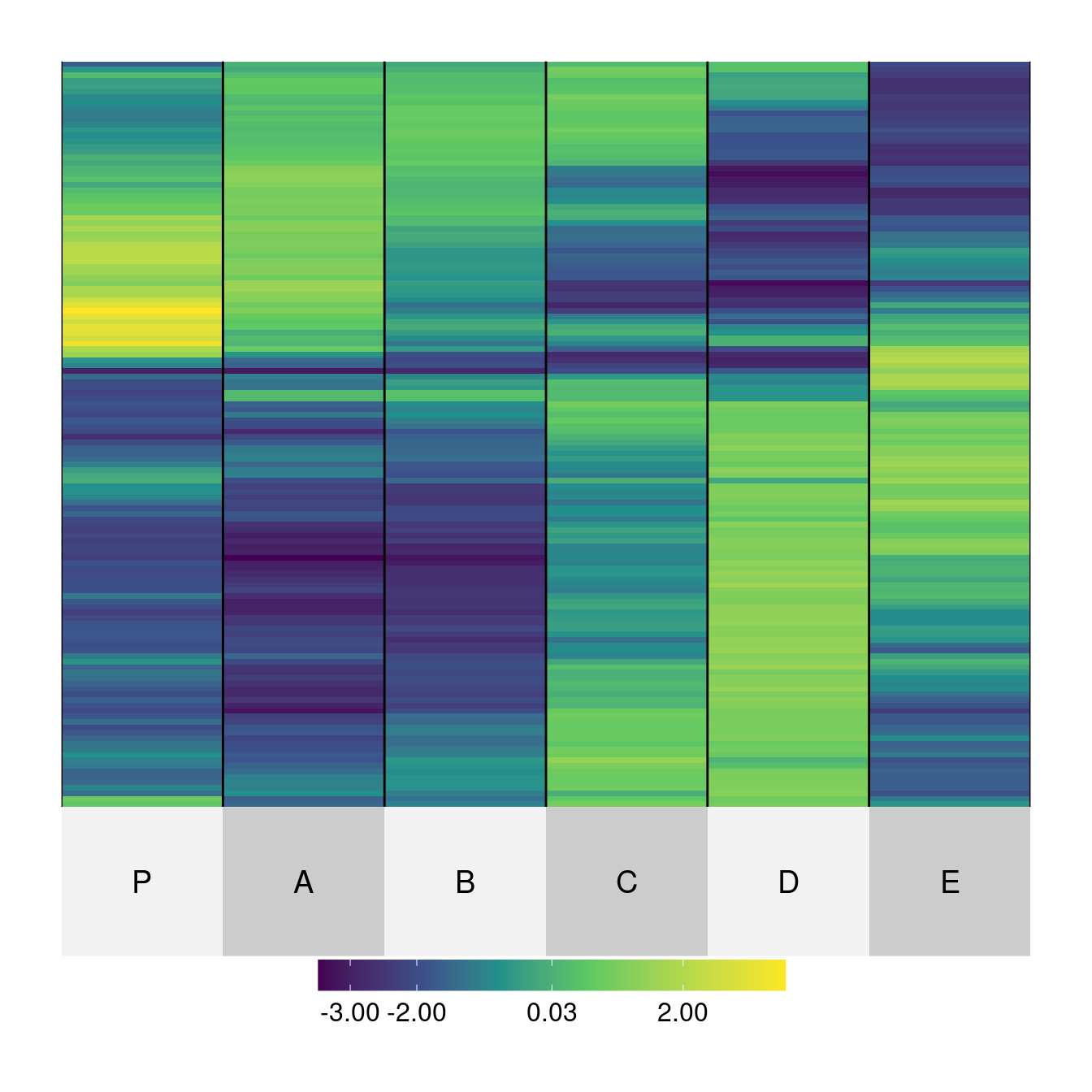
The package creates interactive heatmaps. It already uses package seriation for parameter seriate and supports the methods OLO () and GW.
if (!suppressMessages(require("heatmaply"))) install.packages("heatmaply")
library("heatmaply")
heatmaply(Wood, seriate = "none", main = "HC")
heatmaply(Wood, seriate = "OLO", main = "OLO")Any dendrogram-based seriation method from seriation can be supplied.
o <- seriate(Wood, method = "Heatmap", seriation_method = "OLO_ward")
heatmaply(Wood, Rowv = o[[1]], Colv = o[[2]], main = "OLO (Ward)")
o <- seriate(Wood, method = "Heatmap", seriation_method = "Spectral")
heatmaply(Wood, Rowv = get_rank(o, 1), Colv = get_rank(o, 2), main = "Spectral")