3 Classification: Basic Concepts and Techniques
This chapter introduces decision trees for classification and discusses how models are built and evaluated.
The corresponding chapter of the data mining textbook is available online: Chapter 3: Classification: Basic Concepts and Techniques.
Packages Used in this Chapter
pkgs <- c("caret", "FSelector", "lattice", "mlbench",
"palmerpenguins", "party", "pROC", "rpart", "rpart.plot",
"sampling", "tidyverse")
pkgs_install <- pkgs[!(pkgs %in% installed.packages()[,"Package"])]
if(length(pkgs_install)) install.packages(pkgs_install)The packages used for this chapter are: caret (M. Kuhn 2023), FSelector (Romanski, Kotthoff, and Schratz 2023), lattice (Sarkar 2023), mlbench (Leisch and Dimitriadou 2024), palmerpenguins (Horst, Hill, and Gorman 2022), party (Hothorn et al. 2024), pROC (Robin et al. 2023), rpart (Therneau and Atkinson 2023), rpart.plot (Milborrow 2024), sampling (Tillé and Matei 2023), tidyverse (Wickham 2023c)
In the examples in this book, we use the popular machine learning R package caret. It makes preparing
training sets, building classification (and regression) models and
evaluation easier. A great cheat sheet can be found
here.
A newer R framework for machine learning
is tidymodels, a set of packages that
integrate more naturally with tidyverse. Using tidymodels, or any other
framework (e.g., Python’s scikit-learn) should
be relatively easy after
learning the concepts using caret.
3.1 Basic Concepts
Classification is a machine learning task with the goal to learn a predictive function of the form
\[y = f(\mathbf{x}),\]
where \(\mathbf{x}\) is called the attribute set and \(y\) the class label. The attribute set consists of feature which describe an object. These features can be measured using any scale (i.e., nominal, interval, …). The class label is a nominal attribute. It it is a binary attribute, then the problem is called a binary classification problem.
Classification learns the classification model from training data where both the features and the correct class label are available. This is why it is called a supervised learning problem.
A related supervised learning problem is regression, where \(y\) is a number instead of a label. Linear regression is a very popular supervised learning model which is taught in almost any introductory statistics course. Code examples for regression are available in the extra Chapter Regression*.
This chapter will introduce decision trees, model evaluation and comparison, feature selection, and then explore methods to handle the class imbalance problem.
You can read the free sample chapter from the textbook (Tan, Steinbach, and Kumar 2005): Chapter 3. Classification: Basic Concepts and Techniques
3.2 General Framework for Classification
Supervised learning has two steps:
- Induction: Training a model on training data with known class labels.
- Deduction: Predicting class labels for new data.
We often test model by predicting the class for data where we know the correct label. We test the model on test data with known labels and can then calculate the error by comparing the prediction with the known correct label. It is tempting to measure how well the model has learned the training data, by testing it on the training data. The error on the training data is called resubstitution error. It does not help us to find out if the model generalizes well to new data that was not part of the training.
We typically want to evaluate how well the model generalizes new data, so it is important that the test data and the training data do not overlap. We call the error on proper test data the generalization error.
This chapter builds up the needed concepts. A complete example of how to perform model selection and estimate the generalization error is in the section Hyperparameter Tuning.
3.2.1 The Zoo Dataset
To demonstrate classification, we will use the Zoo dataset which is included in the R package mlbench (you may have to install it). The Zoo dataset containing 17 (mostly logical) variables for 101 animals as a data frame with 17 columns (hair, feathers, eggs, milk, airborne, aquatic, predator, toothed, backbone, breathes, venomous, fins, legs, tail, domestic, catsize, type). The first 16 columns represent the feature vector \(\mathbf{x}\) and the last column called type is the class label \(y\). We convert the data frame into a tidyverse tibble (optional).
data(Zoo, package="mlbench")
head(Zoo)
## hair feathers eggs milk airborne aquatic predator toothed
## aardvark TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE
## antelope TRUE FALSE FALSE TRUE FALSE FALSE FALSE TRUE
## bass FALSE FALSE TRUE FALSE FALSE TRUE TRUE TRUE
## bear TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE
## boar TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE
## buffalo TRUE FALSE FALSE TRUE FALSE FALSE FALSE TRUE
## backbone breathes venomous fins legs tail domestic catsize
## aardvark TRUE TRUE FALSE FALSE 4 FALSE FALSE TRUE
## antelope TRUE TRUE FALSE FALSE 4 TRUE FALSE TRUE
## bass TRUE FALSE FALSE TRUE 0 TRUE FALSE FALSE
## bear TRUE TRUE FALSE FALSE 4 FALSE FALSE TRUE
## boar TRUE TRUE FALSE FALSE 4 TRUE FALSE TRUE
## buffalo TRUE TRUE FALSE FALSE 4 TRUE FALSE TRUE
## type
## aardvark mammal
## antelope mammal
## bass fish
## bear mammal
## boar mammal
## buffalo mammalNote: data.frames in R can have row names. The Zoo data set uses the
animal name as the row names. tibbles from tidyverse do not support
row names. To keep the animal name you can add a column with the animal
name.
library(tidyverse)
as_tibble(Zoo, rownames = "animal")
## # A tibble: 101 × 18
## animal hair feathers eggs milk airborne aquatic predator toothed
## <chr> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl> <lgl>
## 1 aardv… TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE
## 2 antel… TRUE FALSE FALSE TRUE FALSE FALSE FALSE TRUE
## 3 bass FALSE FALSE TRUE FALSE FALSE TRUE TRUE TRUE
## 4 bear TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE
## 5 boar TRUE FALSE FALSE TRUE FALSE FALSE TRUE TRUE
## 6 buffa… TRUE FALSE FALSE TRUE FALSE FALSE FALSE TRUE
## 7 calf TRUE FALSE FALSE TRUE FALSE FALSE FALSE TRUE
## 8 carp FALSE FALSE TRUE FALSE FALSE TRUE FALSE TRUE
## 9 catfi… FALSE FALSE TRUE FALSE FALSE TRUE TRUE TRUE
## 10 cavy TRUE FALSE FALSE TRUE FALSE FALSE FALSE TRUE
## # ℹ 91 more rows
## # ℹ 9 more variables: backbone <lgl>, breathes <lgl>, venomous <lgl>,
## # fins <lgl>, legs <int>, tail <lgl>, domestic <lgl>,
## # catsize <lgl>, type <fct>You will have to remove the animal column before learning a model! In the following I use the data.frame.
I translate all the TRUE/FALSE values into factors (nominal). This is
often needed for building models. Always check summary() to make sure
the data is ready for model learning.
Zoo <- Zoo |>
mutate(across(where(is.logical),
function (x) factor(x, levels = c(TRUE, FALSE)))) |>
mutate(across(where(is.character), factor))
summary(Zoo)
## hair feathers eggs milk airborne aquatic
## TRUE :43 TRUE :20 TRUE :59 TRUE :41 TRUE :24 TRUE :36
## FALSE:58 FALSE:81 FALSE:42 FALSE:60 FALSE:77 FALSE:65
##
##
##
##
##
## predator toothed backbone breathes venomous fins
## TRUE :56 TRUE :61 TRUE :83 TRUE :80 TRUE : 8 TRUE :17
## FALSE:45 FALSE:40 FALSE:18 FALSE:21 FALSE:93 FALSE:84
##
##
##
##
##
## legs tail domestic catsize type
## Min. :0.00 TRUE :75 TRUE :13 TRUE :44 mammal :41
## 1st Qu.:2.00 FALSE:26 FALSE:88 FALSE:57 bird :20
## Median :4.00 reptile : 5
## Mean :2.84 fish :13
## 3rd Qu.:4.00 amphibian : 4
## Max. :8.00 insect : 8
## mollusc.et.al:103.3 Decision Tree Classifiers
We use here the recursive partitioning implementation which follows largely CART and uses the Gini index to make splitting decisions and early stopping (also called pre-pruning).
3.3.1 Create Tree
We create first a tree with the default settings (see ? rpart.control).
tree_default <- Zoo |>
rpart(type ~ ., data = _)
tree_default
## n= 101
##
## node), split, n, loss, yval, (yprob)
## * denotes terminal node
##
## 1) root 101 60 mammal (0.41 0.2 0.05 0.13 0.04 0.079 0.099)
## 2) milk=TRUE 41 0 mammal (1 0 0 0 0 0 0) *
## 3) milk=FALSE 60 40 bird (0 0.33 0.083 0.22 0.067 0.13 0.17)
## 6) feathers=TRUE 20 0 bird (0 1 0 0 0 0 0) *
## 7) feathers=FALSE 40 27 fish (0 0 0.12 0.33 0.1 0.2 0.25)
## 14) fins=TRUE 13 0 fish (0 0 0 1 0 0 0) *
## 15) fins=FALSE 27 17 mollusc.et.al (0 0 0.19 0 0.15 0.3 0.37)
## 30) backbone=TRUE 9 4 reptile (0 0 0.56 0 0.44 0 0) *
## 31) backbone=FALSE 18 8 mollusc.et.al (0 0 0 0 0 0.44 0.56) *Notes:
|>supplies the data forrpart. Sincedatais not the first argument ofrpart, the syntaxdata = _is used to specify where the data inZoogoes. The call is equivalent totree_default <- rpart(type ~ ., data = Zoo).The formula models the
typevariable by all other features represented by..The class variable needs to be a factor to be recognized as nominal or rpart will create a regression tree instead of a decision tree. Use
as.factor()on the column with the class label first, if necessary.
We can plot the resulting decision tree.
library(rpart.plot)
rpart.plot(tree_default, extra = 2)
Note: extra=2 prints for each leaf node the number of correctly
classified objects from data and the total number of objects from the
training data falling into that node (correct/total).
3.3.2 Make Predictions for New Data
I will make up my own animal: A lion with feathered wings.
my_animal <- tibble(hair = TRUE, feathers = TRUE, eggs = FALSE,
milk = TRUE, airborne = TRUE, aquatic = FALSE, predator = TRUE,
toothed = TRUE, backbone = TRUE, breathes = TRUE, venomous = FALSE,
fins = FALSE, legs = 4, tail = TRUE, domestic = FALSE,
catsize = FALSE, type = NA)The data types need to match the original data so we change the columns to be factors like in the training set.
my_animal <- my_animal |>
mutate(across(where(is.logical),
function(x) factor(x, levels = c(TRUE, FALSE))))
my_animal
## # A tibble: 1 × 17
## hair feathers eggs milk airborne aquatic predator toothed
## <fct> <fct> <fct> <fct> <fct> <fct> <fct> <fct>
## 1 TRUE TRUE FALSE TRUE TRUE FALSE TRUE TRUE
## # ℹ 9 more variables: backbone <fct>, breathes <fct>, venomous <fct>,
## # fins <fct>, legs <dbl>, tail <fct>, domestic <fct>,
## # catsize <fct>, type <fct>Next, we make a prediction using the default tree
predict(tree_default , my_animal, type = "class")
## 1
## mammal
## 7 Levels: mammal bird reptile fish amphibian ... mollusc.et.al3.3.3 Manual Calculation of the Resubstitution Error
We will calculate error of the model on the training data manually first, so we see how it is calculated.
predict(tree_default, Zoo) |> head ()
## mammal bird reptile fish amphibian insect mollusc.et.al
## aardvark 1 0 0 0 0 0 0
## antelope 1 0 0 0 0 0 0
## bass 0 0 0 1 0 0 0
## bear 1 0 0 0 0 0 0
## boar 1 0 0 0 0 0 0
## buffalo 1 0 0 0 0 0 0
pred <- predict(tree_default, Zoo, type="class")
head(pred)
## aardvark antelope bass bear boar buffalo
## mammal mammal fish mammal mammal mammal
## 7 Levels: mammal bird reptile fish amphibian ... mollusc.et.alWe can easily tabulate the true and predicted labels to create a confusion matrix.
confusion_table <- with(Zoo, table(type, pred))
confusion_table
## pred
## type mammal bird reptile fish amphibian insect
## mammal 41 0 0 0 0 0
## bird 0 20 0 0 0 0
## reptile 0 0 5 0 0 0
## fish 0 0 0 13 0 0
## amphibian 0 0 4 0 0 0
## insect 0 0 0 0 0 0
## mollusc.et.al 0 0 0 0 0 0
## pred
## type mollusc.et.al
## mammal 0
## bird 0
## reptile 0
## fish 0
## amphibian 0
## insect 8
## mollusc.et.al 10The counts in the diagonal are correct predictions. Off-diagonal counts represent errors (i.e., confusions).
We can summarize the confusion matrix using the accuracy measure.
error <- confusion_table |> sum() - correct
error
## [1] 12
accuracy <- correct / (correct + error)
accuracy
## [1] 0.8812Here is the accuracy calculation as a simple function.
3.3.4 Confusion Matrix using Caret
The caret package provides a convenient way to create and analyze a confusion table including many useful statistics.
library(caret)
confusionMatrix(data = pred,
reference = Zoo |> pull(type))
## Confusion Matrix and Statistics
##
## Reference
## Prediction mammal bird reptile fish amphibian insect
## mammal 41 0 0 0 0 0
## bird 0 20 0 0 0 0
## reptile 0 0 5 0 4 0
## fish 0 0 0 13 0 0
## amphibian 0 0 0 0 0 0
## insect 0 0 0 0 0 0
## mollusc.et.al 0 0 0 0 0 8
## Reference
## Prediction mollusc.et.al
## mammal 0
## bird 0
## reptile 0
## fish 0
## amphibian 0
## insect 0
## mollusc.et.al 10
##
## Overall Statistics
##
## Accuracy : 0.881
## 95% CI : (0.802, 0.937)
## No Information Rate : 0.406
## P-Value [Acc > NIR] : <2e-16
##
## Kappa : 0.843
##
## Mcnemar's Test P-Value : NA
##
## Statistics by Class:
##
## Class: mammal Class: bird Class: reptile
## Sensitivity 1.000 1.000 1.0000
## Specificity 1.000 1.000 0.9583
## Pos Pred Value 1.000 1.000 0.5556
## Neg Pred Value 1.000 1.000 1.0000
## Prevalence 0.406 0.198 0.0495
## Detection Rate 0.406 0.198 0.0495
## Detection Prevalence 0.406 0.198 0.0891
## Balanced Accuracy 1.000 1.000 0.9792
## Class: fish Class: amphibian Class: insect
## Sensitivity 1.000 0.0000 0.0000
## Specificity 1.000 1.0000 1.0000
## Pos Pred Value 1.000 NaN NaN
## Neg Pred Value 1.000 0.9604 0.9208
## Prevalence 0.129 0.0396 0.0792
## Detection Rate 0.129 0.0000 0.0000
## Detection Prevalence 0.129 0.0000 0.0000
## Balanced Accuracy 1.000 0.5000 0.5000
## Class: mollusc.et.al
## Sensitivity 1.000
## Specificity 0.912
## Pos Pred Value 0.556
## Neg Pred Value 1.000
## Prevalence 0.099
## Detection Rate 0.099
## Detection Prevalence 0.178
## Balanced Accuracy 0.956Note: Calculating accuracy on the training data is not a good idea. Keep on reading!
3.4 Model Overfitting
We are tempted to create the largest possible tree
to get the most accurate model. This can be achieved by
changing the algorithms hyperparameter (parameters that
change how the algorithm works). We
set the complexity parameter cp to 0 (split
even if it does not improve the tree) and we set the minimum number of
observations in a node needed to split to the smallest value of 2 (see:
?rpart.control). Note: This is not a good idea!
As we will see later, full trees overfit the training data!
tree_full <- Zoo |>
rpart(type ~ . , data = _,
control = rpart.control(minsplit = 2, cp = 0))
rpart.plot(tree_full, extra = 2,
roundint=FALSE,
box.palette = list("Gy", "Gn", "Bu", "Bn",
"Or", "Rd", "Pu")) # specify 7 colors
tree_full
## n= 101
##
## node), split, n, loss, yval, (yprob)
## * denotes terminal node
##
## 1) root 101 60 mammal (0.41 0.2 0.05 0.13 0.04 0.079 0.099)
## 2) milk=TRUE 41 0 mammal (1 0 0 0 0 0 0) *
## 3) milk=FALSE 60 40 bird (0 0.33 0.083 0.22 0.067 0.13 0.17)
## 6) feathers=TRUE 20 0 bird (0 1 0 0 0 0 0) *
## 7) feathers=FALSE 40 27 fish (0 0 0.12 0.33 0.1 0.2 0.25)
## 14) fins=TRUE 13 0 fish (0 0 0 1 0 0 0) *
## 15) fins=FALSE 27 17 mollusc.et.al (0 0 0.19 0 0.15 0.3 0.37)
## 30) backbone=TRUE 9 4 reptile (0 0 0.56 0 0.44 0 0)
## 60) aquatic=FALSE 4 0 reptile (0 0 1 0 0 0 0) *
## 61) aquatic=TRUE 5 1 amphibian (0 0 0.2 0 0.8 0 0)
## 122) eggs=FALSE 1 0 reptile (0 0 1 0 0 0 0) *
## 123) eggs=TRUE 4 0 amphibian (0 0 0 0 1 0 0) *
## 31) backbone=FALSE 18 8 mollusc.et.al (0 0 0 0 0 0.44 0.56)
## 62) airborne=TRUE 6 0 insect (0 0 0 0 0 1 0) *
## 63) airborne=FALSE 12 2 mollusc.et.al (0 0 0 0 0 0.17 0.83)
## 126) predator=FALSE 4 2 insect (0 0 0 0 0 0.5 0.5)
## 252) legs>=3 2 0 insect (0 0 0 0 0 1 0) *
## 253) legs< 3 2 0 mollusc.et.al (0 0 0 0 0 0 1) *
## 127) predator=TRUE 8 0 mollusc.et.al (0 0 0 0 0 0 1) *Error on the training set of the full tree
pred_full <- predict(tree_full, Zoo, type = "class")
accuracy(Zoo |> pull(type), pred_full)
## [1] 1We see that the error is smaller then for the pruned tree. This, however, does not mean that the model is better. It actually is overfitting the training data (it just memorizes it) and has worse generalization performance on new data. This effect is called overfitting the training data and needs to be avoided.
3.5 Model Selection
We often can create many different models for a classification problem. Above, we have created a decision tree using the default settings and also a full tree. The question is: Which one should we use. This problem is called model selection.
In order to select the model we need to split the training data into a validation set and the training set that is actually used to train model. The error rate on the validation set can then be used to choose between several models.
Caret has model selection build into the train() function. We will select
between the default complexity cp = 0.01 and a full tree cp = 0 (see tuneGrid).
trControl specified how the validation set is obtained. LGOCV
picks randomly the proportion p of data to train and uses the rest as
the validation set. To get a better estimate of the
error, this process is repeated number times and the errors are averaged.
fit <- Zoo |>
train(type ~ .,
data = _ ,
method = "rpart",
control = rpart.control(minsplit = 2), # we have little data
tuneGrid = data.frame(cp = c(0.01, 0)),
trControl = trainControl(method = "LGOCV", p = 0.8, number = 10),
tuneLength = 5)
fit
## CART
##
## 101 samples
## 16 predictor
## 7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
##
## No pre-processing
## Resampling: Repeated Train/Test Splits Estimated (10 reps, 80%)
## Summary of sample sizes: 83, 83, 83, 83, 83, 83, ...
## Resampling results across tuning parameters:
##
## cp Accuracy Kappa
## 0.00 0.9444 0.9232
## 0.01 0.9389 0.9155
##
## Accuracy was used to select the optimal model using the
## largest value.
## The final value used for the model was cp = 0.We see that in this case, the full tree model performs slightly better. This is a result of the fact that we only have a very small dataset.
3.6 Model Evaluation
Models should be evaluated on a test set that has no overlap with the training set. We typically split the data using random sampling. To get reproducible results, we set random number generator seed.
set.seed(2000)3.6.1 Holdout Method
Test data is not used in the model building process and set aside purely for testing the model. Here, we partition data the 80% training and 20% testing.
inTrain <- createDataPartition(y = Zoo$type, p = .8)[[1]]
Zoo_train <- Zoo |> slice(inTrain)
Zoo_test <- Zoo |> slice(-inTrain)Now we can train on the test set and get the generalization error on the test set.
3.6.2 Cross-Validation
k-fold cross-validation splits the data randomly into \(k\) folds. It then holds one fold back for testing and trains on the other \(k-1\) folds. This is done with each fold and the resulting statistic (e.g., accuracy) is averaged. This method uses the data more efficiently then the holdout method.
Cross validation can be directly used in train() using
trControl = trainControl(method = "cv", number = 10).
If no model selection is necessary then this will give the
generalization error.
Cross-validation runs are independent and can be done faster in
parallel. To enable multi-core support, caret uses the package
foreach and you need to load a do backend. For Linux, you can use
doMC with 4 cores. Windows needs different backend like doParallel
(see caret cheat sheet above).
## Linux backend
# library(doMC)
# registerDoMC(cores = 4)
# getDoParWorkers()
## Windows backend
# library(doParallel)
# cl <- makeCluster(4, type="SOCK")
# registerDoParallel(cl)3.7 Hyperparameter Tuning
Hyperparameters are parameters that change how a
training algorithm works. An example is the complexity parameter
cp for rpart decision trees. Tuning the hyperparameter means that
we want to perform model selection to pick the best setting.
We typically use the holdout method to create a test set and then use cross validation using the training data for model selection. Let us use 80% for training and hold out 20% for testing.
inTrain <- createDataPartition(y = Zoo$type, p = .8)[[1]]
Zoo_train <- Zoo |> slice(inTrain)
Zoo_test <- Zoo |> slice(-inTrain)The package caret combines training and validation for hyperparameter
tuning into the train() function. It internally splits the
data into training and validation sets and thus will provide you with
error estimates for different hyperparameter settings. trainControl is
used to choose how testing is performed.
For rpart, train tries to tune the cp parameter (tree complexity)
using accuracy to chose the best model. I set minsplit to 2 since we
have not much data. Note: Parameters used for tuning (in this case
cp) need to be set using a data.frame in the argument tuneGrid!
Setting it in control will be ignored.
fit <- Zoo_train |>
train(type ~ .,
data = _ ,
method = "rpart",
control = rpart.control(minsplit = 2), # we have little data
trControl = trainControl(method = "cv", number = 10),
tuneLength = 5)
fit
## CART
##
## 83 samples
## 16 predictors
## 7 classes: 'mammal', 'bird', 'reptile', 'fish', 'amphibian', 'insect', 'mollusc.et.al'
##
## No pre-processing
## Resampling: Cross-Validated (10 fold)
## Summary of sample sizes: 74, 73, 77, 75, 73, 75, ...
## Resampling results across tuning parameters:
##
## cp Accuracy Kappa
## 0.00 0.9289 0.9058
## 0.08 0.8603 0.8179
## 0.16 0.7296 0.6422
## 0.22 0.6644 0.5448
## 0.32 0.4383 0.1136
##
## Accuracy was used to select the optimal model using the
## largest value.
## The final value used for the model was cp = 0.Note: Train has built 10 trees using the training folds for each
value of cp and the reported values for accuracy and Kappa are the
averages on the validation folds.
A model using the best tuning parameters and using all the data supplied
to train() is available as fit$finalModel.
library(rpart.plot)
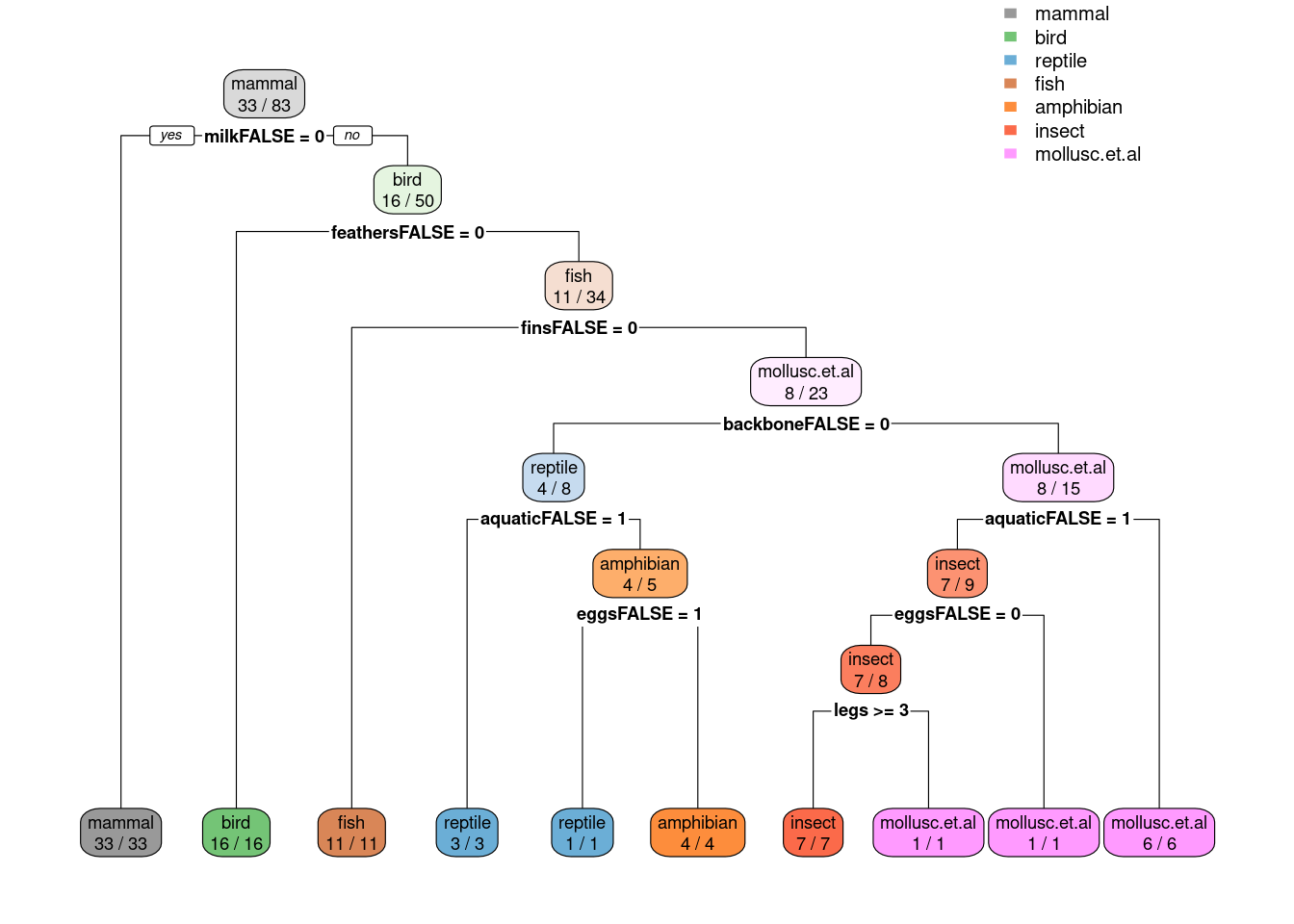
rpart.plot(fit$finalModel, extra = 2,
box.palette = list("Gy", "Gn", "Bu", "Bn", "Or", "Rd", "Pu"))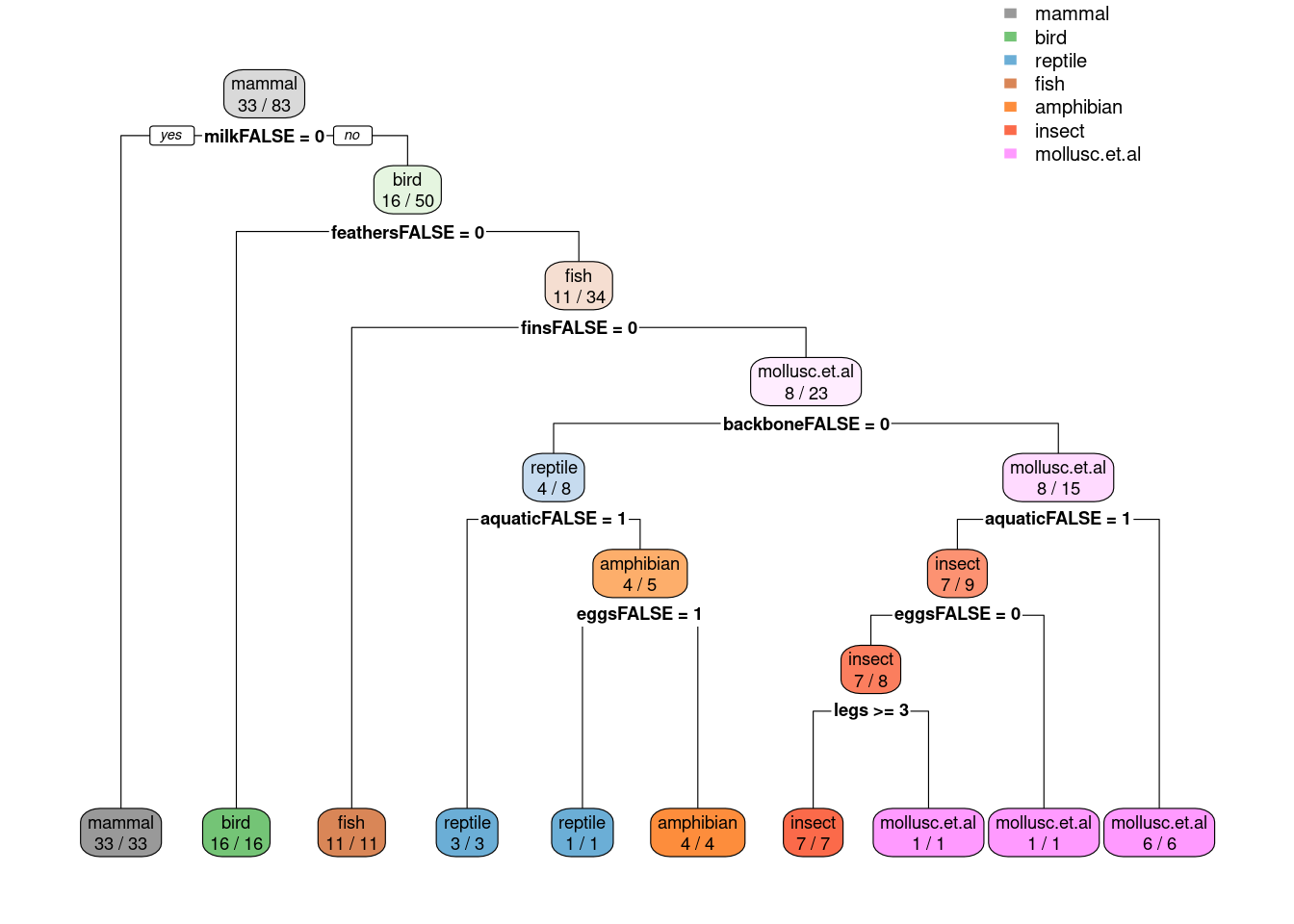
caret also computes variable importance. By default it uses competing
splits (splits which would be runners up, but do not get chosen by the
tree) for rpart models (see ? varImp). Toothed is the runner up for
many splits, but it never gets chosen!
varImp(fit)
## rpart variable importance
##
## Overall
## toothedFALSE 100.0
## feathersFALSE 79.5
## eggsFALSE 67.7
## milkFALSE 63.3
## backboneFALSE 57.3
## finsFALSE 53.5
## hairFALSE 52.1
## breathesFALSE 48.9
## legs 41.4
## tailFALSE 29.0
## aquaticFALSE 27.5
## airborneFALSE 26.5
## predatorFALSE 10.6
## venomousFALSE 1.8
## catsizeFALSE 0.0
## domesticFALSE 0.0Here is the variable importance without competing splits.
imp <- varImp(fit, compete = FALSE)
imp
## rpart variable importance
##
## Overall
## milkFALSE 100.00
## feathersFALSE 55.69
## finsFALSE 39.45
## aquaticFALSE 28.11
## backboneFALSE 21.76
## eggsFALSE 12.32
## legs 7.28
## tailFALSE 0.00
## domesticFALSE 0.00
## airborneFALSE 0.00
## catsizeFALSE 0.00
## toothedFALSE 0.00
## venomousFALSE 0.00
## hairFALSE 0.00
## breathesFALSE 0.00
## predatorFALSE 0.00
ggplot(imp)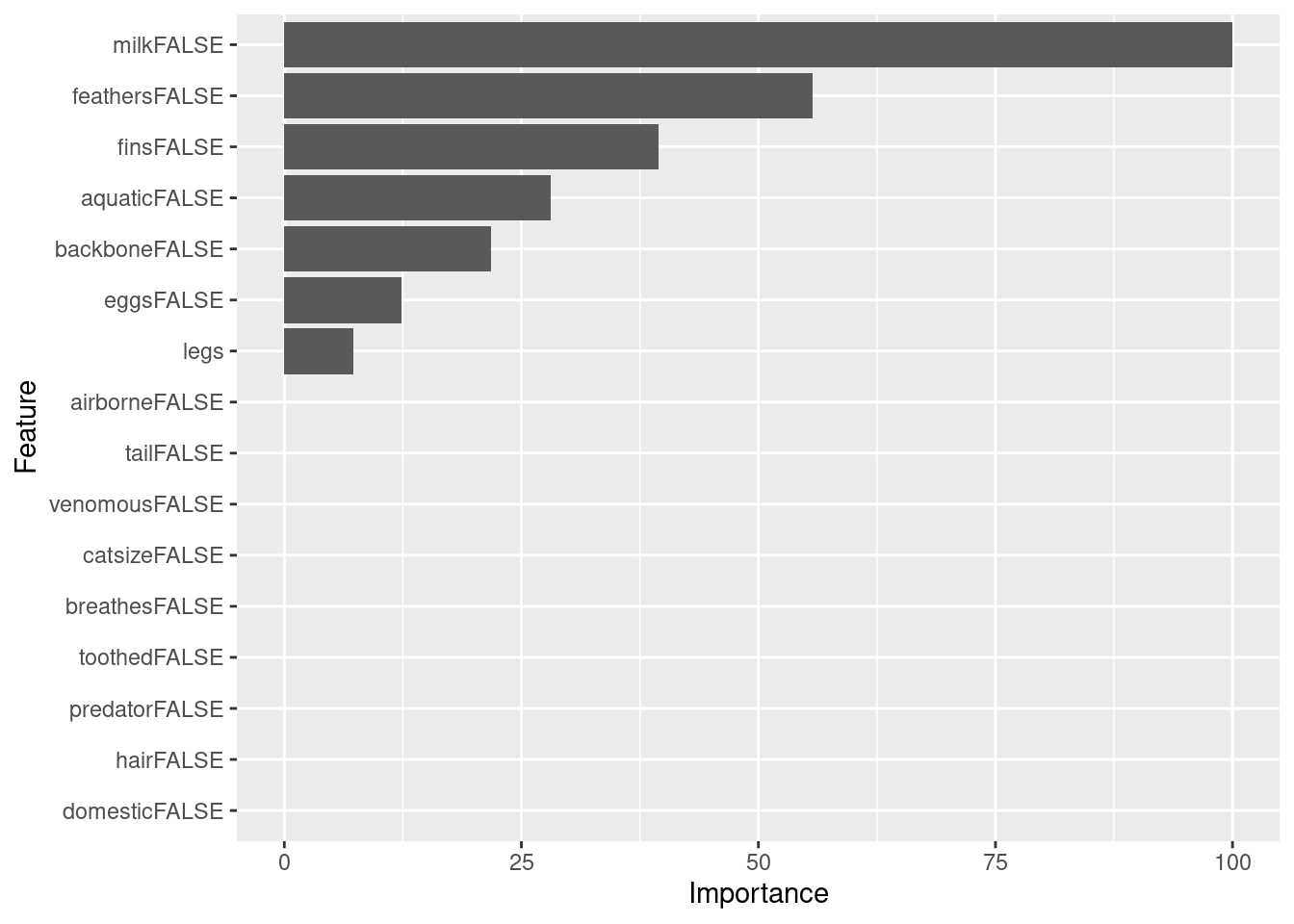
Note: Not all models provide a variable importance function. In this
case caret might calculate the variable importance by itself and ignore
the model (see ? varImp)!
Now, we can estimate the generalization error of the best model on the held out test data.
pred <- predict(fit, newdata = Zoo_test)
pred
## [1] mammal bird mollusc.et.al bird
## [5] mammal mammal insect bird
## [9] mammal mammal mammal mammal
## [13] bird fish fish reptile
## [17] mammal mollusc.et.al
## 7 Levels: mammal bird reptile fish amphibian ... mollusc.et.alCaret’s confusionMatrix() function calculates accuracy, confidence
intervals, kappa and many more evaluation metrics. You need to use
separate test data to create a confusion matrix based on the
generalization error.
confusionMatrix(data = pred,
ref = Zoo_test |> pull(type))
## Confusion Matrix and Statistics
##
## Reference
## Prediction mammal bird reptile fish amphibian insect
## mammal 8 0 0 0 0 0
## bird 0 4 0 0 0 0
## reptile 0 0 1 0 0 0
## fish 0 0 0 2 0 0
## amphibian 0 0 0 0 0 0
## insect 0 0 0 0 0 1
## mollusc.et.al 0 0 0 0 0 0
## Reference
## Prediction mollusc.et.al
## mammal 0
## bird 0
## reptile 0
## fish 0
## amphibian 0
## insect 0
## mollusc.et.al 2
##
## Overall Statistics
##
## Accuracy : 1
## 95% CI : (0.815, 1)
## No Information Rate : 0.444
## P-Value [Acc > NIR] : 4.58e-07
##
## Kappa : 1
##
## Mcnemar's Test P-Value : NA
##
## Statistics by Class:
##
## Class: mammal Class: bird Class: reptile
## Sensitivity 1.000 1.000 1.0000
## Specificity 1.000 1.000 1.0000
## Pos Pred Value 1.000 1.000 1.0000
## Neg Pred Value 1.000 1.000 1.0000
## Prevalence 0.444 0.222 0.0556
## Detection Rate 0.444 0.222 0.0556
## Detection Prevalence 0.444 0.222 0.0556
## Balanced Accuracy 1.000 1.000 1.0000
## Class: fish Class: amphibian Class: insect
## Sensitivity 1.000 NA 1.0000
## Specificity 1.000 1 1.0000
## Pos Pred Value 1.000 NA 1.0000
## Neg Pred Value 1.000 NA 1.0000
## Prevalence 0.111 0 0.0556
## Detection Rate 0.111 0 0.0556
## Detection Prevalence 0.111 0 0.0556
## Balanced Accuracy 1.000 NA 1.0000
## Class: mollusc.et.al
## Sensitivity 1.000
## Specificity 1.000
## Pos Pred Value 1.000
## Neg Pred Value 1.000
## Prevalence 0.111
## Detection Rate 0.111
## Detection Prevalence 0.111
## Balanced Accuracy 1.000Some notes
- Many classification algorithms and
trainin caret do not deal well with missing values. If your classification model can deal with missing values (e.g.,rpart) then usena.action = na.passwhen you calltrainandpredict. Otherwise, you need to remove observations with missing values withna.omitor use imputation to replace the missing values before you train the model. Make sure that you still have enough observations left. - Make sure that nominal variables (this includes logical variables) are coded as factors.
- The class variable for train in caret cannot have level names that
are keywords in R (e.g.,
TRUEandFALSE). Rename them to, for example, “yes” and “no.” - Make sure that nominal variables (factors) have examples for all
possible values. Some methods might have problems with variable
values without examples. You can drop empty levels using
droplevelsorfactor. - Sampling in train might create a sample that does not contain examples for all values in a nominal (factor) variable. You will get an error message. This most likely happens for variables which have one very rare value. You may have to remove the variable.
3.8 Pitfalls of Model Selection and Evaluation
- The training data and the test sets cannot overlap or we will not evaluate the generalization performance. The training set can be come contaminated by things like preprocessing the all the data together.
- Do not measure the error on the training set or use the validation error as a generalization error estimate. Always use the generalization error on a test set!
3.9 Model Comparison
We will compare decision trees with a k-nearest neighbors (kNN)
classifier. We will create fixed sampling scheme (10-folds) so we
compare the different models using exactly the same folds. It is
specified as trControl during training.
train_index <- createFolds(Zoo_train$type, k = 10, returnTrain = TRUE)Build models
rpartFit <- Zoo_train |>
train(type ~ .,
data = _,
method = "rpart",
tuneLength = 10,
trControl = trainControl(method = "cv", index = train_index)
)Note: for kNN we ask train to scale the data using
preProcess = "scale". Logicals will be used as 0-1 variables in
Euclidean distance calculation.
knnFit <- Zoo_train |>
train(type ~ .,
data = _,
method = "knn",
preProcess = "scale",
tuneLength = 10,
trControl = trainControl(method = "cv", index = train_index)
)Compare accuracy over all folds.
resamps <- resamples(list(
CART = rpartFit,
kNearestNeighbors = knnFit
))
summary(resamps)
##
## Call:
## summary.resamples(object = resamps)
##
## Models: CART, kNearestNeighbors
## Number of resamples: 10
##
## Accuracy
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## CART 0.6667 0.750 0.7778 0.7890 0.8512 0.875 0
## kNearestNeighbors 0.7500 0.875 0.8990 0.9076 1.0000 1.000 0
##
## Kappa
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## CART 0.5909 0.6800 0.7188 0.7227 0.7706 0.8298 0
## kNearestNeighbors 0.6800 0.8315 0.8713 0.8816 1.0000 1.0000 0caret provides some visualizations. For
example, a boxplot to compare the accuracy and kappa distribution (over
the 10 folds).
We see that kNN is performing consistently better on the folds than CART (except for some outlier folds).
Find out if one models is statistically better than the other (is the difference in accuracy is not zero).
difs <- diff(resamps)
difs
##
## Call:
## diff.resamples(x = resamps)
##
## Models: CART, kNearestNeighbors
## Metrics: Accuracy, Kappa
## Number of differences: 1
## p-value adjustment: bonferroni
summary(difs)
##
## Call:
## summary.diff.resamples(object = difs)
##
## p-value adjustment: bonferroni
## Upper diagonal: estimates of the difference
## Lower diagonal: p-value for H0: difference = 0
##
## Accuracy
## CART kNearestNeighbors
## CART -0.119
## kNearestNeighbors 0.00768
##
## Kappa
## CART kNearestNeighbors
## CART -0.159
## kNearestNeighbors 0.00538p-values tells you the probability of seeing an even more extreme value
(difference between accuracy) given that the null hypothesis (difference
= 0) is true. For a better classifier, the p-value should be less than
.05 or 0.01. diff automatically applies Bonferroni correction for
multiple comparisons. In this case, kNN seems better but the classifiers
do not perform statistically differently.
3.10 Feature Selection*
Decision trees implicitly select features for splitting, but we can also select features before we apply any learning algorithm. Since different features lead to different models, choosing the best set of features is also a type of model selection.
Many feature selection methods are implemented in the FSelector package.
3.10.1 Univariate Feature Importance Score
These scores measure how related each feature is to the class variable. For discrete features (as in our case), the chi-square statistic can be used to derive a score.
weights <- Zoo_train |>
chi.squared(type ~ ., data = _) |>
as_tibble(rownames = "feature") |>
arrange(desc(attr_importance))
weights
## # A tibble: 16 × 2
## feature attr_importance
## <chr> <dbl>
## 1 feathers 1
## 2 milk 1
## 3 backbone 1
## 4 toothed 0.981
## 5 eggs 0.959
## 6 breathes 0.917
## 7 hair 0.906
## 8 fins 0.845
## 9 legs 0.834
## 10 airborne 0.818
## 11 tail 0.779
## 12 aquatic 0.725
## 13 catsize 0.602
## 14 venomous 0.520
## 15 predator 0.374
## 16 domestic 0.256We can plot the importance in descending order (using reorder to order factor
levels used by ggplot).
ggplot(weights,
aes(x = attr_importance, y = reorder(feature, attr_importance))) +
geom_bar(stat = "identity") +
xlab("Importance score") +
ylab("Feature")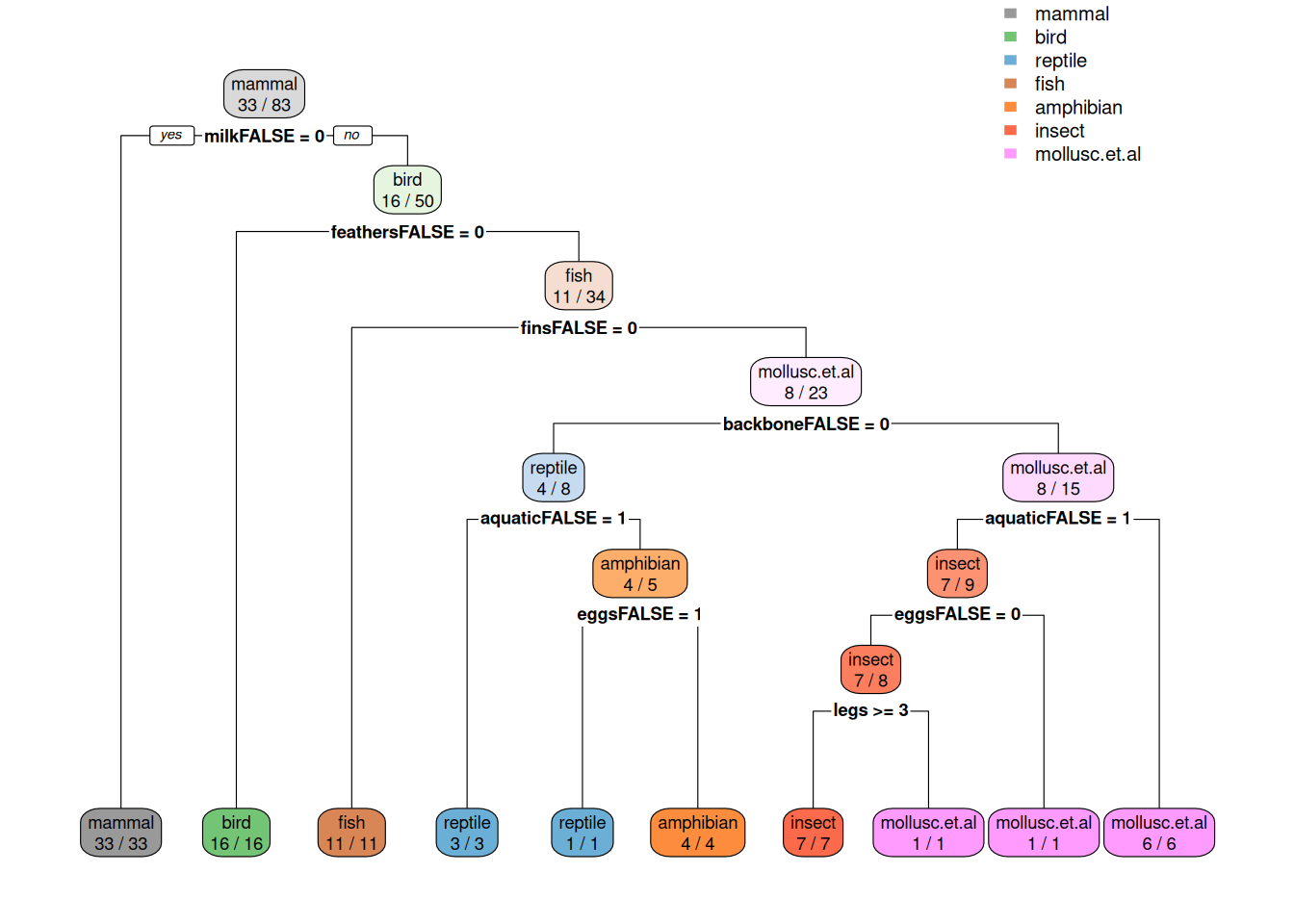
Picking the best features is called the feature ranking approach. Here we pick the 5 highest-ranked features.
subset <- cutoff.k(weights |>
column_to_rownames("feature"), 5)
subset
## [1] "feathers" "milk" "backbone" "toothed" "eggs"Use only the selected features to build a model (Fselector provides
as.simple.formula).
f <- as.simple.formula(subset, "type")
f
## type ~ feathers + milk + backbone + toothed + eggs
## <environment: 0x5aa5331a7d78>
m <- Zoo_train |> rpart(f, data = _)
rpart.plot(m, extra = 2, roundint = FALSE)
There are many alternative ways to calculate univariate importance scores (see package FSelector). Some of them (also) work for continuous features. One example is the information gain ratio based on entropy as used in decision tree induction.
Zoo_train |>
gain.ratio(type ~ ., data = _) |>
as_tibble(rownames = "feature") |>
arrange(desc(attr_importance))
## # A tibble: 16 × 2
## feature attr_importance
## <chr> <dbl>
## 1 milk 1
## 2 backbone 1
## 3 feathers 1
## 4 toothed 0.959
## 5 eggs 0.907
## 6 breathes 0.845
## 7 hair 0.781
## 8 fins 0.689
## 9 legs 0.689
## 10 airborne 0.633
## 11 tail 0.573
## 12 aquatic 0.474
## 13 venomous 0.429
## 14 catsize 0.310
## 15 domestic 0.115
## 16 predator 0.1103.10.2 Feature Subset Selection
Often, features are related and calculating importance for each feature
independently is not optimal. We can use greedy search heuristics. For
example cfs uses correlation/entropy with best first search.
Zoo_train |>
cfs(type ~ ., data = _)
## [1] "hair" "feathers" "eggs" "milk" "toothed" "backbone"
## [7] "breathes" "fins" "legs" "tail"Black-box feature selection uses an evaluator function (the black box)
to calculate a score to be maximized.
This is typically the best method, since it can use the actual model
for selection.
First, we define an evaluation
function that builds a model given a subset of features and calculates a
quality score. We use here the average for 5 bootstrap samples
(method = "cv" can also be used instead), no tuning (to be faster),
and the average accuracy as the score.
evaluator <- function(subset) {
model <- Zoo_train |>
train(as.simple.formula(subset, "type"),
data = _,
method = "rpart",
trControl = trainControl(method = "boot", number = 5),
tuneLength = 0)
results <- model$resample$Accuracy
cat("Trying features:", paste(subset, collapse = " + "), "\n")
m <- mean(results)
cat("Accuracy:", round(m, 2), "\n\n")
m
}Start with all features (but not the class variable type)
There are several (greedy) search strategies available. These run for a while so we have commented them out. Remove the comment for one at a time to try this type of feature selection.
#subset <- backward.search(features, evaluator)
#subset <- forward.search(features, evaluator)
#subset <- best.first.search(features, evaluator)
#subset <- hill.climbing.search(features, evaluator)
#
#subset3.10.3 Using Dummy Variables for Factors
Nominal features (factors) are often encoded as a series of 0-1 dummy variables. For example, let us try to predict if an animal is a predator given the type. First we use the original encoding of type as a factor with several values.
tree_predator <- Zoo_train |>
rpart(predator ~ type, data = _)
rpart.plot(tree_predator, extra = 2, roundint = FALSE)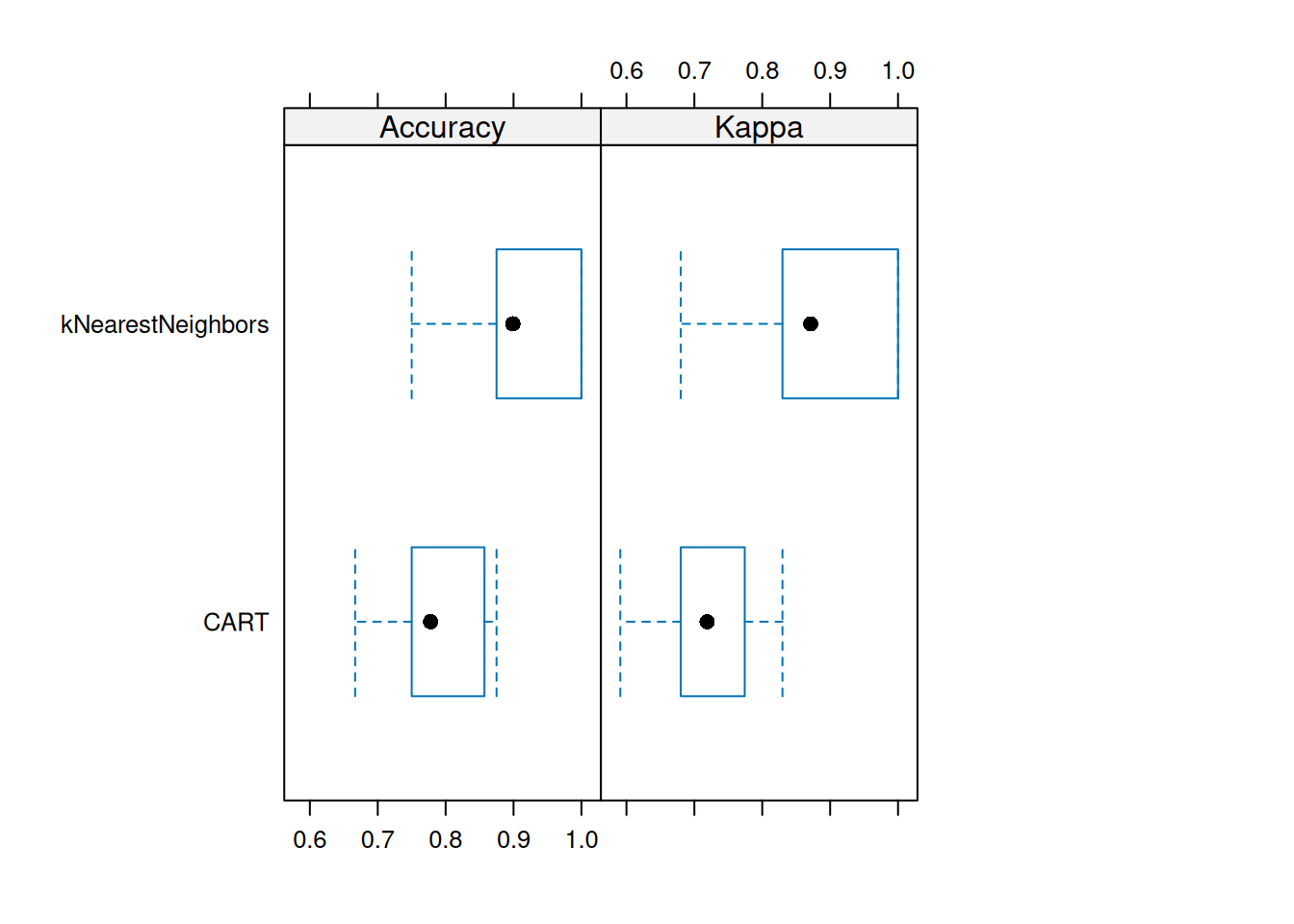
Note: Some splits use multiple values. Building the tree will become extremely slow if a factor has many levels (different values) since the tree has to check all possible splits into two subsets. This situation should be avoided.
Convert type into a set of 0-1 dummy variables using class2ind. See
also ? dummyVars in package caret.
Zoo_train_dummy <- as_tibble(class2ind(Zoo_train$type)) |>
mutate(across(everything(), as.factor)) |>
add_column(predator = Zoo_train$predator)
Zoo_train_dummy
## # A tibble: 83 × 8
## mammal bird reptile fish amphibian insect mollusc.et.al predator
## <fct> <fct> <fct> <fct> <fct> <fct> <fct> <fct>
## 1 1 0 0 0 0 0 0 TRUE
## 2 0 0 0 1 0 0 0 TRUE
## 3 1 0 0 0 0 0 0 TRUE
## 4 1 0 0 0 0 0 0 TRUE
## 5 1 0 0 0 0 0 0 FALSE
## 6 1 0 0 0 0 0 0 FALSE
## 7 0 0 0 1 0 0 0 FALSE
## 8 0 0 0 1 0 0 0 TRUE
## 9 1 0 0 0 0 0 0 FALSE
## 10 1 0 0 0 0 0 0 TRUE
## # ℹ 73 more rows
tree_predator <- Zoo_train_dummy |>
rpart(predator ~ .,
data = _,
control = rpart.control(minsplit = 2, cp = 0.01))
rpart.plot(tree_predator, roundint = FALSE)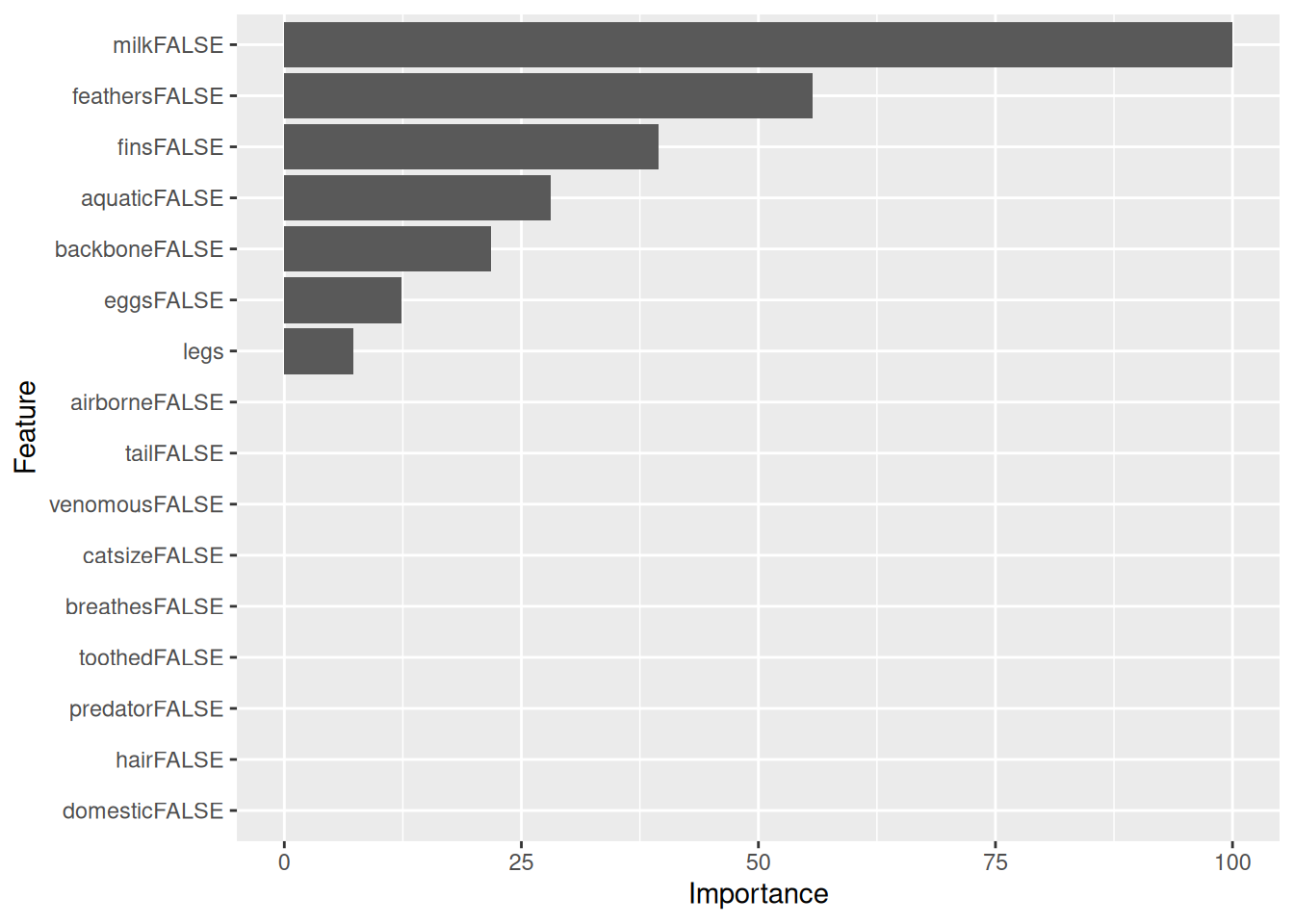
Using caret on the original factor encoding automatically translates
factors (here type) into 0-1 dummy variables (e.g., typeinsect = 0).
The reason is that some models cannot directly use factors and caret
tries to consistently work with all of them.
fit <- Zoo_train |>
train(predator ~ type,
data = _,
method = "rpart",
control = rpart.control(minsplit = 2),
tuneGrid = data.frame(cp = 0.01))
fit
## CART
##
## 83 samples
## 1 predictor
## 2 classes: 'TRUE', 'FALSE'
##
## No pre-processing
## Resampling: Bootstrapped (25 reps)
## Summary of sample sizes: 83, 83, 83, 83, 83, 83, ...
## Resampling results:
##
## Accuracy Kappa
## 0.5635 0.1166
##
## Tuning parameter 'cp' was held constant at a value of 0.01
rpart.plot(fit$finalModel, extra = 2)
Note: To use a fixed value for the tuning parameter cp, we have to
create a tuning grid that only contains that value.
3.11 Exercises*
We will use again the Palmer penguin data for the exercises.
library(palmerpenguins)
head(penguins)
## # A tibble: 6 × 8
## species island bill_length_mm bill_depth_mm flipper_length_mm
## <chr> <chr> <dbl> <dbl> <dbl>
## 1 Adelie Torgersen 39.1 18.7 181
## 2 Adelie Torgersen 39.5 17.4 186
## 3 Adelie Torgersen 40.3 18 195
## 4 Adelie Torgersen NA NA NA
## 5 Adelie Torgersen 36.7 19.3 193
## 6 Adelie Torgersen 39.3 20.6 190
## # ℹ 3 more variables: body_mass_g <dbl>, sex <chr>, year <dbl>Create a R markdown file with the code and discussion for the following below. Remember, the complete approach is described in section Hyperparameter Tuning.
- Split the data into a training and test set.
- Create an rpart decision tree to predict the species. You will have to deal with missing values.
- Experiment with setting
minsplitfor rpart and make suretuneLengthis at least 5. Discuss the model selection process (hyperparameter tuning) and what final model was chosen. - Visualize the tree and discuss what the splits mean.
- Calculate the variable importance from the fitted model. What variables are the most important? What variables do not matter?
- Use the test set to evaluate the generalization error and accuracy.